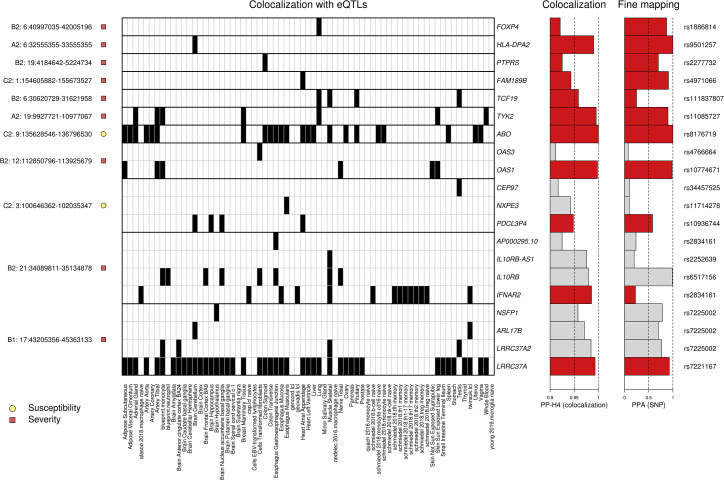
Figure 3.
Colocalization of COVID-19 GWAS signals and eQTLs
Heatmap showing tissue eQTLs that colocalized with GWAS signals in 11 loci. Shown are (1) on the left, each locus and its association with susceptibility or severity; (2) in the middle, a heatmap showing the posterior probability of association (PP-H4) between eQTLs in the indicated tissue and the GWAS signal in each locus (black = PP-H4 ≥ 0.9); (3) on the right, barplots indicating the PP-H4 of the indicated eQTL and GWAS signals and of the lead variant tested for each colocalization. “PP-H4” indicates the PP of the GWAS and eQTL signals to have the same underlying causal variant: since all associations shown have PP-H4 > 0.9, the x-axis scale is from 0.9 to 1. PPA (SNP) indicates the PPA for the indicated SNP to be causal for each colocalization. The corresponding RS ID is shown on the right. For each of the four loci with eQTL signals for more than one gene, and only the associations for the gene with the highest PP-H4 (indicated by red bars) were used to fine map the locus. The colocalization data are shown for one phenotype (C2, A1, B1, or B2) per locus and are indicated on the left before the chromosome number and locus coordinates. At one severity-associated locus (1:154605882-155673527), we found that FAM189B colocalized with high PPA (PPA = 0.94) only with the C2 phenotype, but not with any of the severity phenotypes (PPA < 0.67; Table S2).