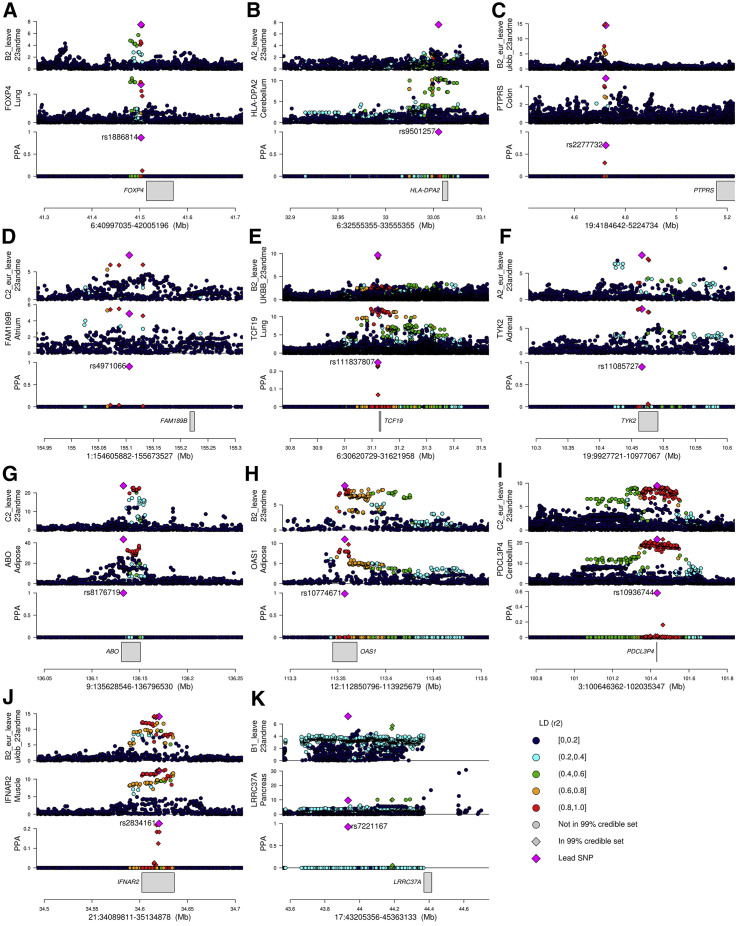
Figure 4.
Colocalizations between COVID-19 GWAS and tissue eQTL signals
For each of the 11 fine-mapped loci, four panels are presented: (1) GWAS p values for indicated COVID-19 phenotype; (2) eQTL p values for indicated gene; (3) posterior probability of colocalization of each variant (coloc PP; calculated using coloc) and color-coded based on linkage disequilibrium in individuals of European descent (r2) calculated using LDlinkR (Myers et al., 2020); and (4) location of gene in locus. The p values of all variants tested in the GWAS and eQTL studies are shown; however, only variants tested in both studies were used for colocalization.
(A) Locus 6:40997035-42005196, phenotype B2 GWAS (B2_ALL_leave_23andme), and FOXP4 eQTL in lung.
(B) Locus 6:32555355-33555355, phenotype A2 GWAS (A2_ALL_leave_23andme), and HLA-DPA2 eQTL in brain cerebellum.
(C) Locus 19:4184642-5224734, phenotype B2 GWAS (B2_ALL_eur_leave_ukbb_23andme), and PTPRS eQTL in colon sigmoid.
(D) Locus 1:154605882-155673527, phenotype C2 GWAS (C2_ALL_eur_leave_23andme), and FAM189B eQTL in heart atrial appendage; although this signal is severity associated, only the C2 phenotype colocalized with an eQTL.
(E) Locus 6:30620729-31621958, phenotype B2 GWAS (B2_ALL_leave_UKBB_23andme), and TCF19 eQTL in lung.
(F) Locus 19:9927721-10977067, phenotype A2 GWAS (A2_ALL_eur_leave_23andme), and TYK2 eQTL in adrenal gland.
(G) Locus 9:135628546-136796530, phenotype C2 GWAS (C2_ALL_leave_23andme), and ABO eQTL in adipose visceral omentum.
(H) Locus 12:112850796-113925679, phenotype B2 GWAS (B2_ALL_leave_23andme), and OAS1 eQTL in adipose subcutaneous.
(I) Locus 3:100646362-102035347, phenotype C2 GWAS (C2_ALL_eur_leave_23andme), and PDCL3P4 eQTL in cerebellum; since the FOXP4 locus was associated with non-European individuals, LD is shown for East Asians.
(J) Locus 21:34089811-35134878, phenotype B2 GWAS (B2_ALL_eur_leave_ukbb_23andme), and IFNAR2 eQTL in muscle skeletal.
(K) Locus 17:43205356-45363133, phenotype B1 GWAS (B1_ALL_leave_23andme), and LRRC37A eQTL in pancreas.