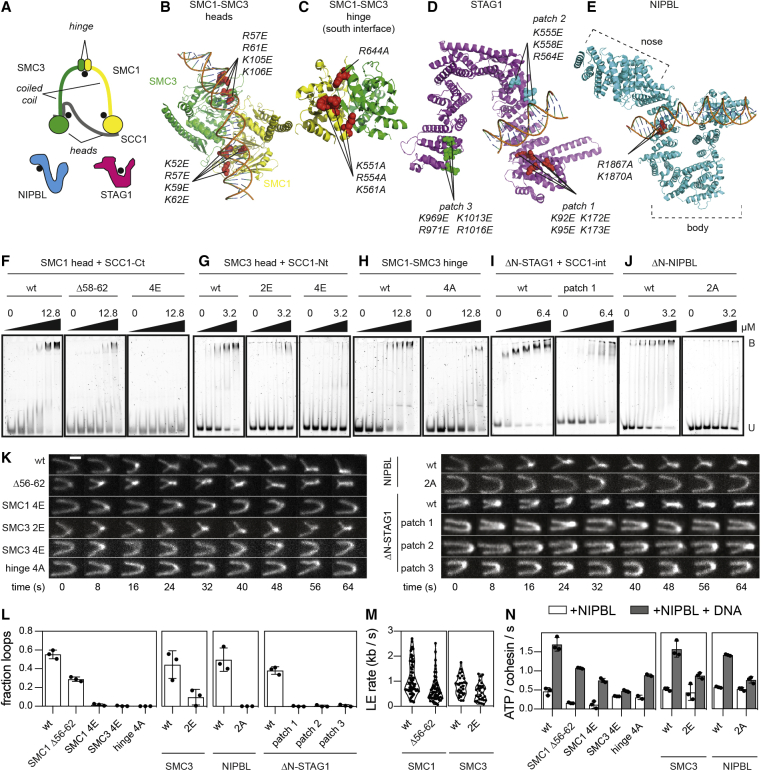
Figure 1.
DNA binding sites on cohesin-NIPBL required for loop extrusion
(A) Cartoon illustration of human cohesin-NIPBL. DNA binding sites are indicated with black dots.
(B–E) Structures of the SMC1-SMC3 ATPase heads (B, PDB: 6WG3), the SMC1-SMC3 hinge (C, PDB: 2WD5), STAG1 (D; PDB: 6WG3), and NIPBL (E; PDB: 6WG3). Mutated residues are indicated as spheres.
(F–J) Electrophoretic mobility shift assays (EMSAs) of the SMC1 head (F), the SMC3 head (G), the hinge (H), ΔN-STAG1 (I), and ΔN-NIPBL (J). Increasing concentrations of wild-type (wt) or mutant proteins were incubated with a 40 base pair double stranded DNA and separated by native PAGE. U, unbound DNA; B, bound DNA.
(K) Time-lapse recordings of loop extrusion events catalyzed by wt and mutant cohesin complexes. DNA was stained with Sytox Orange. Scale bar, 2 μm.
(L) Frequencies of loop extrusion events. Shown are means ± SD and values of individual replicates.
(M) Rates of loop extrusion in kilobases per second (kb/s). Shown are rates of individual loop extrusion events and a violin plot of their distribution.
(N) NIPBL- and DNA-stimulated ATPase rates of the indicated cohesin complexes. Hydrolysis of ATP was detected by thin layer chromatography. Shown are means ± SD and values of individual replicates.
See also Figure S1.