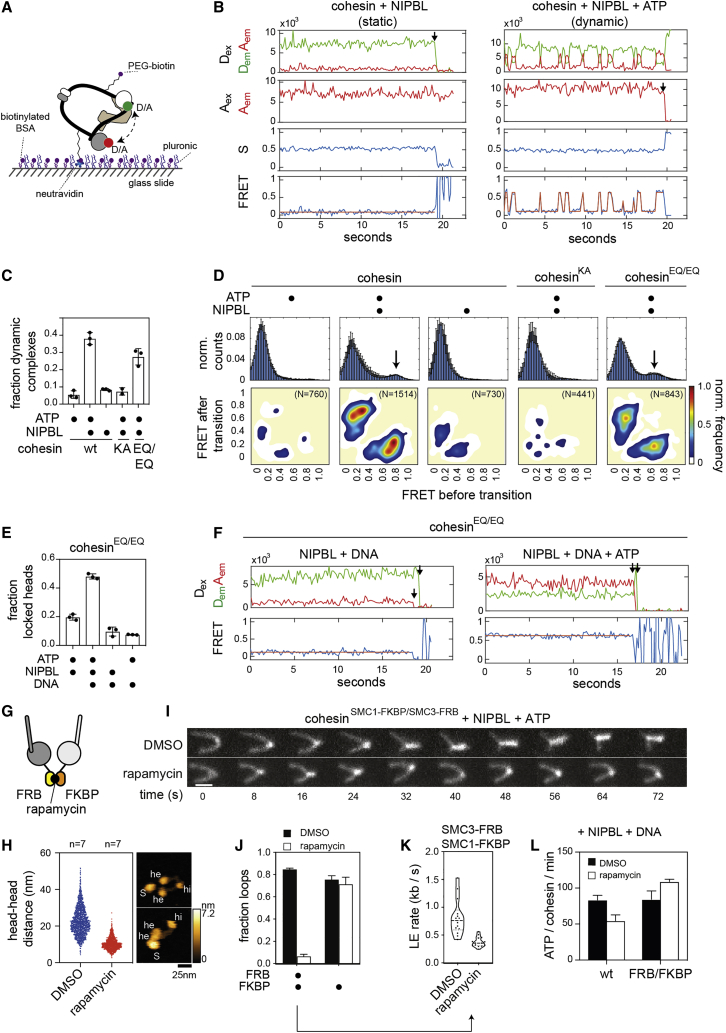
Figure 3.
Dynamics of SMC head engagements visualized by smFRET
(A) Experimental setup of single molecule FRET (smFRET) experiments.
(B) Example smFRET traces of head engagement. Dex, direct donor excitation; Aex, direct acceptor excitation; S, stoichiometry; FRET, FRET efficiency; Dem, Aem, donor and acceptor emission in artificial units (A.U.); HMM, hidden Markov model fits of the raw FRET traces. Black arrows indicate donor or acceptor bleaching.
(C) Fraction of wild-type (wt) and mutant cohesin molecules undergoing dynamic head engagements. Shown are means ± SD and values of individual replicates.
(D) Normalized FRET distributions (top row) and transition density plots (bottom row) of head engagements of different cohesin complexes. N, total number of molecules quantified. The FRET distributions show means ± SD of 3 replicates, with exception of cohesinKA (2 replicates).
(E) Fraction of cohesinEQ/EQ complexes with stably engaged ATPase heads (“locked heads”). Shown are means ± SD and values of individual replicates.
(F) Example traces of disengaged ATP-heads (left) and locked ATPase heads (right).
(G) Cartoon illustration of cohesinSMC1-FKBP/SMC3-FRB.
(H) Distribution of head distances in HS-AFM recordings of cohesinSMC1-FKBP/SMC3-FRB. Shown are distances measured in individual frames. n, number of molecules analyzed. Example stills of cohesinSMC1-FKBP/SMC3-FRB are shown on the right. Heads (he), hinges (hi), and STAG1 (S) are indicated.
(I) Example stills of loop extrusion events by cohesinSMC1-FKBP/SMC3-FRB in presence of DMSO or rapamycin. DNA was stained with Sytox Orange. Scale bar, 2 μm.
(J) Frequencies of loop extrusion events catalyzed by cohesinSMC1-FKBP/SMC3-FRB or cohesinSMC1-FKBP. Shown are means ± SD (3 replicates).
(K) Rates of individual loop extrusion events catalyzed by cohesinSMC1-FKBP/SMC3-FRB and violin plot of their distribution.
(L) ATPase rates of wt cohesin or cohesinSMC1-FKBP/SMC3-FRB. Shown are means ± SD (2 replicates). ATP hydrolysis was determined by thin layer chromatography.
See also Figure S3.