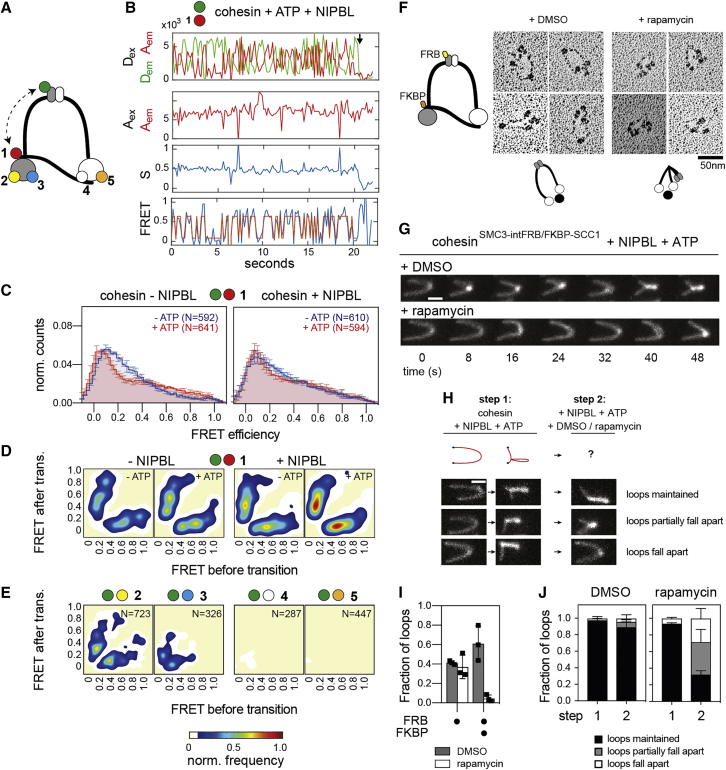
Figure 5.
Dynamics of hinge bending visualized by smFRET
(A) Design of the hinge bending sensors.
(B) Example smFRET trace of sensor pair 1.
(C) FRET distributions of hinge bending (sensor pair 1). Shown are means ± SD (3 replicates). N, number of molecules analyzed.
(D) Transition density plots of sensor pair 1.
(E) Transition density plots of sensor pairs 2–5. Pooled data from 3 replicates (sensors 2 and 3) and 2 replicates (sensors 4 and 5).
(F) Rotary shadowed electron micrographs of cohesinFKBP-SCC1/SMC3-intFRB. Top cartoon: design of cohesinFKBP-SCC1/SMC3-intFRB. Bottom cartoons: observed conformations.
(G) Time-lapse recordings of loop extrusion events by cohesinFKBP-SCC1/SMC3-intFRB in presence of DMSO or rapamycin. DNA was stained with Sytox Orange. Scale bar, 2 μm.
(H) Loop maintenance in presence of DMSO or rapamycin. Loops were first formed by cohesinFKBP-SCC1/SMC3-intFRB (step 1) followed by flow-in of DMSO or rapamycin (step 2). Bottom: example stills of the three major fates of loops upon rapamycin flow-in. Scale bar, 2 μm.
(I) Loop extrusion frequencies of cohesinSMC3-intFRB and cohesinFKBP-SCC1/SMC3-intFRB. Shown are means ± SD and values of individual replicates.
(J) Fraction of loops formed before (1) and after (2) flow-in of DMSO or rapamycin. Shown are means ± SD (3 replicates).
See also Figure S5.