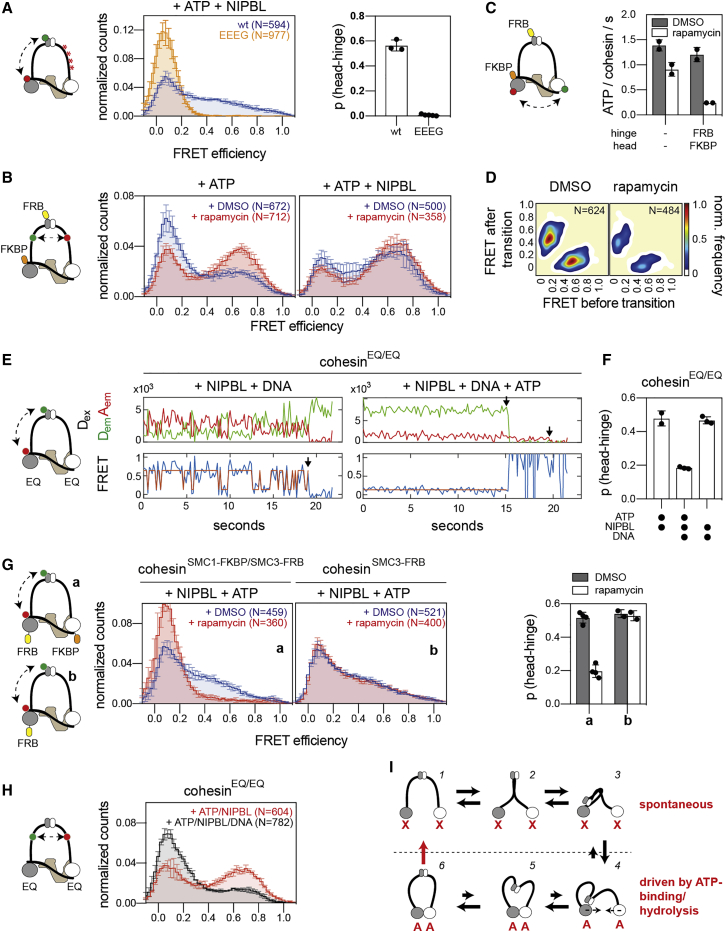
Figure 6.
Coupling of head engagement, coiled coil alignment, and hinge bending
(A) Hinge bending in cohesinEEEG. Left: FRET distributions of wt cohesin (blue, 3 replicates) and cohesinEEEG (orange, 5 replicates). N, total number of molecules. Shown are means ± SD. Right: quantification of the probability of hinge bending. Shown are means ± SD and values of individual replicates.
(B) FRET distributions of coiled coil alignment in cohesinFKBP-SCC1/SMC3-intFRB. Shown are means ± SD (3 replicates).
(C) ATPase activity of cohesinFKBP-SCC1/SMC3-intFRB. Shown are means ± SD and individual replicates. ATP hydrolysis was measured by thin layer chromatography.
(D) Frequencies of head-engagements of cohesinFKBP-SCC1/SMC3-intFRB. Pooled data of 4 replicates per condition.
(E) Example smFRET traces of hinge bending in cohesinEQ/EQ.
(F) Quantification of hinge bending of cohesinEQ/EQ. Shown are means ± SD and values of individual replicates.
(G) FRET distribution and probabilities of hinge bending in cohesinSMC1-FKBP/SMC3-FRB (a) and cohesinSMC1-FKBP (b). Shown are means ± SD and values of individual replicates.
(H) FRET distributions of coiled coil alignment in cohesinEQ/EQ. Shown are means ± SD (3 replicates).
(I) Model of cohesin’s conformational cycle. NIPBL is omitted for clarity. In absence of ATP (“X”), cohesin cycles between separated and aligned states (1 to 2). When aligned, cohesin can cycle between bent and outstretched conformations (2 to 3). Binding of ATP (“A”) promotes head-engagement and coiled coil separation (4). These events promote the displacement of the hinge from the SMC3 head (4 to 6). Upon full head engagement, ATP hydrolysis separates the heads and resets the cycle (6 to 1).
See also Figure S6.