Figure 7.
Nucleotide dependent interface switching of NIPBL
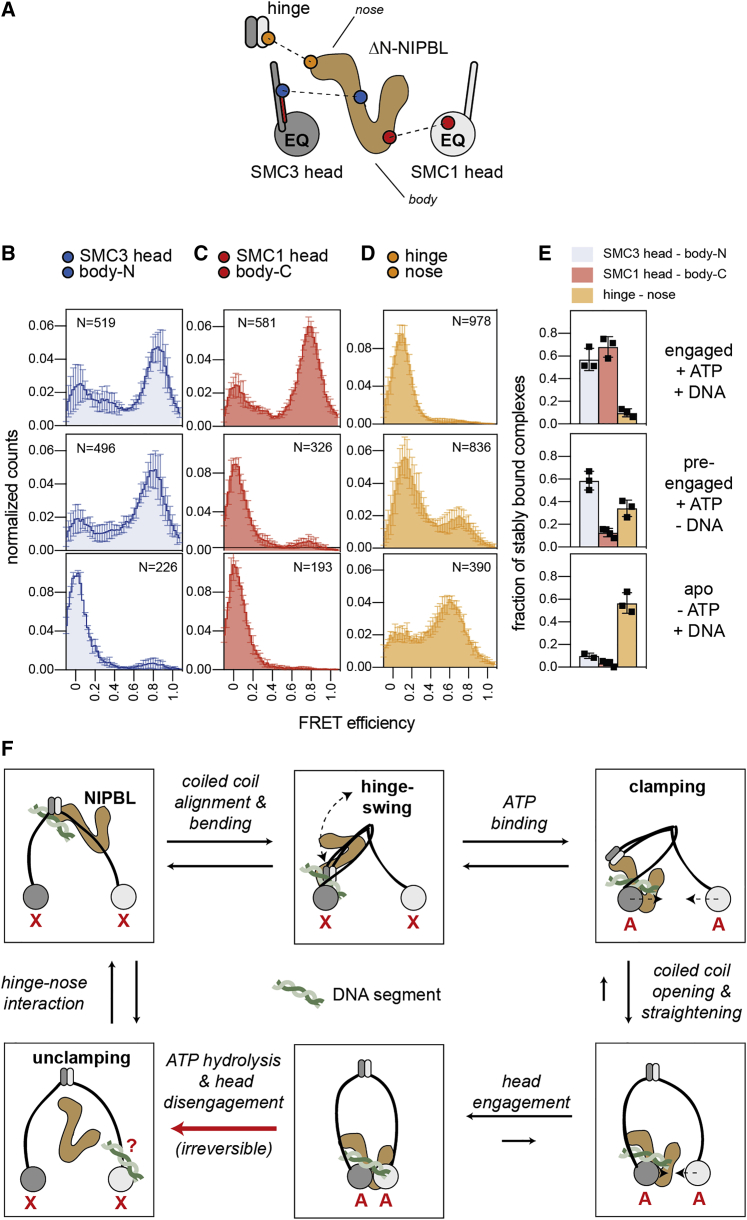
(A) FRET sensor pairs for NIPBL-head and NIPBL-hinge interactions. Body and nose of NIPBL are indicated.
(B) FRET distributions of the SMC3 head-body-N sensor in the engaged, the pre-engaged, and the apo states (top to bottom). N, number of molecules. Shown are means ± SD (3 replicates).
(C) As in (B) but for the SMC1 head-body-C sensor. Shown are means ± SD (3 replicates).
(D) As in (B) but for the hinge-nose sensor. Shown are means ± SD (3 replicates).
(E) Fraction of stably bound complexes. Shown are means ± SD and values of individual replicates.
(D) “Swing and clamp” model of DNA translocation. In the nucleotide free state (“X”), NIPBL interacts via its nose with the hinge. Bending moves the hinge together with NIPBL to the SMC3-head. Upon ATP-binding (“A”), the body of NIPBL forms a clamp with the SMC3 head, inducing the engagement of the heads. The hinge dissociates from the nose and the coiled coils stretch. The engagement of the heads triggers ATP hydrolysis and results in dissociation of the clamp and disengagement of the heads, resetting cohesin for the next cycle. The proposed pathway of a DNA stretch from the hinge to the SMC3 head is indicated. At the end of one cycle, the DNA segment may be transferred to the SMC1 head.
See also Figure S7.