Figure S1.
DNA binding sites in cohesin-NIPBL, related to Figure 1
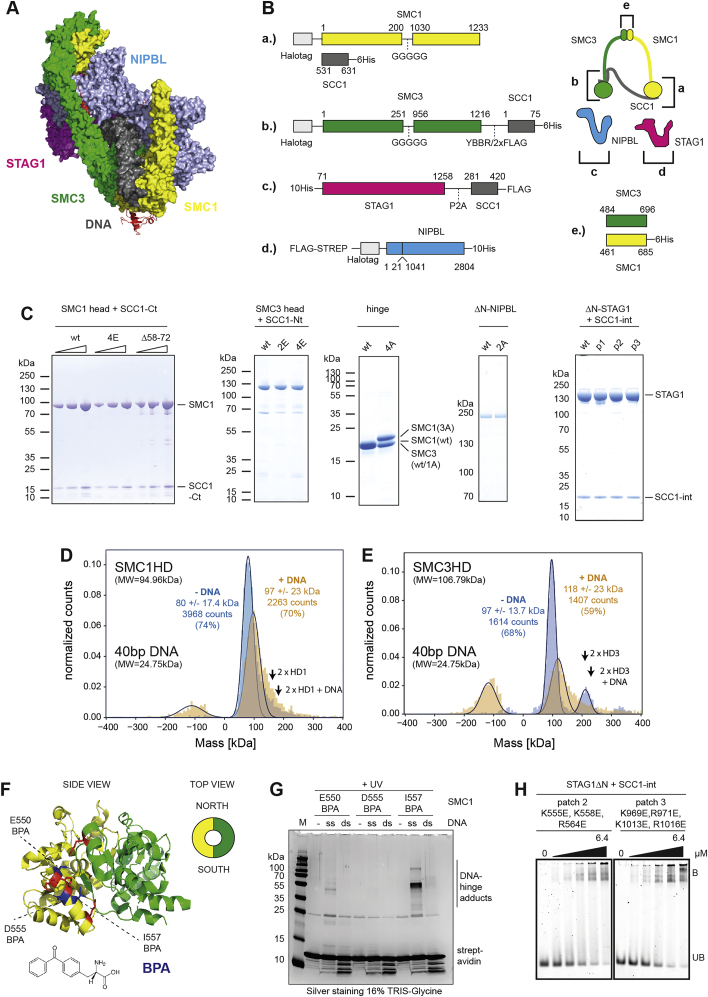
A.) Structure of the DNA clamp formed by the SMC1-SMC3 heads and NIPBL. Shown is a space filling model of PDB: 6WG3.
B.) Design of the SMC1 head domain (a.), SMC3 head domain (b.), ΔN−STAG1 (c.), ΔN−NIPBL (d.) and the hinge domain (e.). With exception of ΔN−NIPBL, constructs were expressed with fragments of SCC1 to facilitate purification. Numbers correspond to the amino acid positions in the respective wild-type (wt) proteins. The cartoon illustration on the top right indicates the location of the individual constructs within the cohesin complex.
C.) Purity of the SMC1 head domain, SMC3 head domain, hinge, ΔN−STAG1, ΔN−NIPBL. Proteins were separated by SDS-PAGE and stained by Coomassie.
D.) Mass photometry analysis of the SMC1 head in presence of ATP and in absence or presence of tenfold molar excess of a 40 base pair (bp) double stranded (ds) DNA. Negative masses correspond to events, where protein-DNA complexes had dissociated from the surface between two frames. The theoretical masses of the SMC1 head and 40 bp DNA are indicated. The expected masses of SMC1 head dimers and complexes of SMC1 head dimers and one DNA molecule are indicated with arrows (“2xHD1” and “2xHD1 + DNA,” respectively). Note that the molecular mass of the 40 bp DNA is below the detection limit of mass photometry. For this reason, DNA alone is not detected.
E.) As in D.), but with the SMC3 head. Note that a small fraction of the SMC3 head preparation contains dimeric head domains, which are a result of mispairing of the fused SCC1 fragments. The expected masses of SMC3 head dimers and complexes between SMC3 head dimers and one DNA molecule are indicated with arrows (“2xHD3” and “2xHD3 + DNA,” respectively).
F.) Structure of the hinge domain (PDB: 2WD5), as viewed from the side. Residues substituted by para-benzoyl-phenylalanine (BPA) via amber suppression are indicated in blue. Residues mutated to alanine in hinge4A are shown in red. The cartoon shows the definition of the “South” and “North” interfaces of the hinge, as viewed from the top. The organic structure of BPA is indicated.
J.) Photo crosslinking of a 40 base single (ss) or double stranded (ds) DNA to the South interface of the hinge. Hinge domains with the indicated residues replaced by BPA were incubated in presence or absence of biotinylated DNA and treated with UV light. DNA was immobilized on streptavidin beads and washed under denaturing conditions. After heat-elution, DNA-hinge adducts were visualized by SDS-PAGE and silver staining. Note that crosslinking residues (550, 557) face toward the South interface whereas the non-crosslinking residue (555) faces away from it. Similar to what has been observed before (Shi et al., 2020), we found higher cross-linking efficiencies for ssDNA compared to dsDNA, possibly due to the increased flexibility of ssDNA at this length, but there is also crosslinking with dsDNA, visible as a smear.
K.) EMSAs of STAG1patch2 and STAG1patch3, as described in Figure 1I.