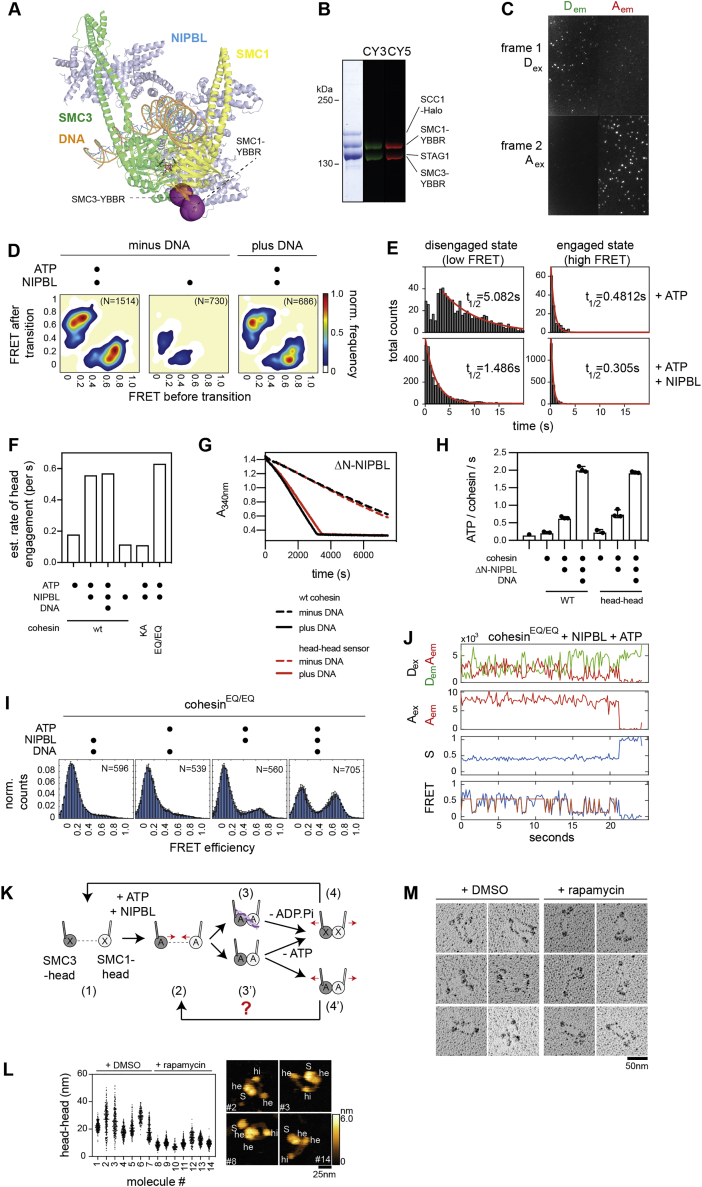
Figure S3.
Kinetics of head engagement visualized by smFRET, related to Figure 3
A.) Positions of the head sensor probes in the cryo-EM model of human cohesin-NIPBL (PDB: 6WG3). The positions of the YBBR-tags are indicated with purple spheres.
B.) Purity and labeling of cohesin with donor and acceptor fluorophores (sulfo-Cy3 and sulfo-Cy5, respectively). CY3: donor fluorescence. CY5: acceptor fluorescence. Molecular weights are indicated in kilo Daltons (kDa). Note that the YBBR-tags are labeled randomly with donor and acceptor molecules.
C.) Example field of view of FRET-labeled cohesin molecules immobilized on glass slides and imaged by TIRF microscopy and alternating laser excitation. Top: First frame of a movie imaged upon direct donor excitation (Dex). Bottom: Second frame imaged upon direct acceptor excitation (Aex). Left: Image of donor emission (Dem). Right: Image of acceptor emission (Aem).
D.) Effect of DNA on the rates of head engagement. Shown are transition density plots of cohesin-NIPBL in presence of ATP and ds DNA. Transition density plots from the two conditions without DNA were taken from Figure 3D for direct comparison. N, total number of molecules.
E.) Dwell time distributions of low and high FRET states (left and right columns, respectively) of cohesin in presence of ATP alone or ATP and NIPBL (top and bottom, respectively). Single exponential decay fits are indicated with a red line. The half time (t1/2) of each fit is shown.
F.) Estimated head engagement rates under different experimental conditions. Engagement rates were quantified as the inverse sum of the half times of the low and high FRET states (as determined in E.).
G.) Kinetics of ATP hydrolysis of wild-type and head sensor cohesin complexes, as measured by a coupled ATPase assay. The assay measures the conversion NADH to NAD by a decrease in the absorbance at 340nm (A340nm), which is coupled to the regeneration of ATP hydrolyzed by cohesin-NIPBL.
H.) Quantification of ATPase rates as measured in G.) under different conditions. A 75-base ds DNA fragment was used as the DNA substrate. Shown are means ± SD and values of individual replicates.
I.) FRET distributions of the experiments shown in Figure 3E. Shown are means ± SD (3 replicates).
J.) Example smFRET trace of cohesinEQ/EQ undergoing reversible head engagement cycles in presence of NIPBL and ATP. Note that the observation of reversible head engagements by cohesinEQ/EQ indicates that head disengagements can occur without prior ATP-hydrolysis.
K.) Model of the head-engagement/disengagement cycle. In the apo-state (1), the heads are disengaged. In the presence of NIPBL (omitted for clarity), the binding of ATP induces the approach and subsequent engagement of the heads (2). Once engaged, in presence of DNA (3), ATP hydrolysis and release allows disengagement (4). In the absence of DNA (3′), the heads can disengage without ATP-hydrolysis (4’). Note that in this case, it is not clear if head disengagement occurs with or without prior ATP dissociation.
L.) Quantification of head distances in hsAFM recordings of cohesinSMC1-FKBP/SMC3-FRB in presence of DMSO or rapamycin for individual cohesin molecules. Each spot corresponds to one frame. The panel on the right shows example stills of the indicated molecules.
M.) Rotary shadowing EM images of cohesinSMC1-FKBP/SMC3-FRB in the presence of DMSO or rapamycin.