Abstract
Background
Native African men (NAM) experience a disproportionate burden of prostate cancer (PCa) and have higher mortality rates compared to European American men (EAM). While socioeconomic status has been implicated as a driver of this disparity, little is known about the genomic mechanisms and distinct biological pathways that are associated with PCa of native men of African origin.
Methods
To understand biological factors that contribute to this disparity we utilized a total of 406 multi-institutional localized PCa samples, collected by MADCaP biospecimen network and Moffitt Cancer Center/University of Pennsylvania Health science system. We performed comparative genomics and immunohistochemistry to identify the biomarkers that are highly enriched in Native African Men from west Africa and compared them with African American (AAM) and European American Men (EAM). Quantified mRNA expression and Median H scores based on immune reactivity of staining cells, were compared using Mann Whitney test. For gene expression analysis, p-values were further adjusted for multiple comparisons using false discovery rates.
Results
IHC analysis on selected biomarkers showed a consistent association between ERG status and race with 83% of NAM exhibiting tumors that lacked TMPRSS2-ERG translocation (ERGnegative) as compared to AAM (71%) and EAM (52%). A higher proportion of NAM (29%) were also found to be double negative (ERGnegative and PTENLoss) as compared to AAM (6%) and EAM (7%). NAM tumors had significantly higher immunoreactivity (H-score) for PSMA, and EZH2, whereas they have lower H-score for PTEN, MYC, AR, RB and Racemase, (all p < 0.05). Comparative genomics revealed that NAM had significant transcriptomic variability in AR-activity score. In pathways enrichment analysis NAM tumors exhibited the enrichment of pro-inflammatory pathways including cytokine, interleukins, inflammatory response, and NFκB signaling.
Conclusions
Prostate tumors in Native African men are genomically distinct and are characterized by the dysregulation of several biomarkers. Furthermore, these tumors are also highly enriched for the major pro-inflammatory pathways. These distinct biological features may have implications for diagnosis and response to targeted therapy among Black men, globally.
Keywords: Native African Men, Prostate Cancer, Race Disparities, Prostate Genomics
Introduction
Globally men of African descent are known to experience a disproportionate burden of prostate cancer (PCa) and have higher mortality rates as compared to their European counterparts 1–3. PCa is also one of the most prevalent cancer among West African men including those from Ghana and Senegal and is attributed to increased mortality rates reported among native west African men (NAM) 2,4. Similarly, African American men (AAM) are also at the highest risk of PCa related disparities with prevalence rates comparable to that of NAM 5. While AAM and NAM are geographically distinct, a greater proportion of AAM in the US finds their ancestral origin from West African countries including Senegal and Ghana 6,7. Although socioeconomic status (SES) including optimal access to care can partially explain outcome level race disparities, they do not account for the disproportionately higher incidence of PCa among men of African origin 8. Moreover, emerging studies have shown comparable outcomes among AAM and European American (EAM) including those from randomized clinical trials; however, most of these studies do not provide a mechanistic explanation to these findings 9,10. Therefore, understanding the distinct tumor features of NAM may reveal molecular underpinnings of race disparities among men of African origin including AAM.
The genetic contribution to PCa disparity has been well established with the identification of significant racial differences in frequency and expression of various genes and biomarkers in AAM 11–14. However, limited data exist to evaluate such molecular distinctions in the tumors from NAM 1. Additionally, genomic features in the tumors of NAM can reflect the inherent potential for disease progression and metastasis which may aid in developing genomically directed therapies to target advance PCa in men of African Descent 11,12,15. In this report, we utilized a unique dataset of native men with detailed mRNA and immunohistochemical (IHC) information to contrast their molecular characteristics with AAM and EAM in the United States. This study attempts to uncover biological pathways that are enriched in men of African origin which could otherwise not be discovered by studying predominant EAM populations alone.
Materials and Methods
Sample acquisition and centralization
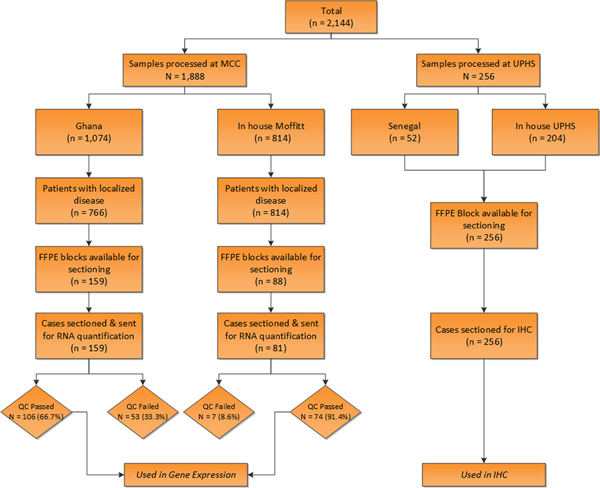
Sample acquisition for this retrospective analysis was undertaken within the Men of African Descent and Carcinoma of the Prostate (MADCaP) biospecimen network which is a pan-African registry of cancer research comprised of recruitment and implementation centers (RICs) in Ghana and Senegal, and monitoring centers in the US 16. Upon approval from institutional review board (IRB) of MADCaP participant sites (National Radiotherapy Oncology and Nuclear Medicine Centre, Korle Bu Teaching Hospital (NRONMC) in Accra, Ghana; Senegal (the Hôpital Général de Grand Yoff [HOGGY]); Moffitt Cancer Center in Florida; and University of Pennsylvania health science system [UPHS] in Pennsylvania), tumor samples were identified from the eligible patients and formalin-fixed paraffin-embedded specimens (FFPE) were selected for subsequent tissue processing. Samples collected from Ghana NRONMC were processed and centralized along with in-house Moffitt cases at MCC, whereas Senegalese samples were centralized along with in-house UPHS cases at UPHS. Both Ghana and Senegalese samples herein are referred to as NAMGhana and NAMSenegal respectively. The consort diagram provides a detailed overview of sample collection and analytical steps employed in the analysis (Fig 1). Tumor samples were centralized by the institutional pathologist to achieve consistent Gleason score grading based on the modified criteria 17. Final analytical dataset consists of total of 408 samples (Fig 1).
Figure 1.
Consort diagram for the samples used in the study.
Gene expression
Targeted gene expression analysis on tumor mRNA was carried out on predominantly biopsy MADCaP samples that were processed at MCC (Ghana n = 106 & EAM n = 46). Unstained sections were obtained and analyzed using the HTG EdgeSeq platform (HTG Molecular Diagnostics, Inc.). Samples were run on an HTG EdgeSeq Processor using the HTG EdgeSeq oncology biomarker panel (OBP) and immuno-oncology (PIP) customized probe sets. A total of 3951 genes were evaluated on relative expression and were compared between race groups. The samples were sequenced on an Illumina MiSeq to generate FASTQ data files. The HTG EdgeSeq Parser was used to align the FASTQ files to the probe sets to collate the data, as previously described. The scaling factor, median gene-level expression value for each sample-gene count adjusted by the geometric mean over all genes, was applied to normalize the data. The final analytical dataset from MCC comprised of samples that passed stringent quality control (QC) metrics for RNA quality and integrity in the HTG EdgeSeq platform. QC failed proportion (n = 60) from both Ghana and MCC tumor samples are provided in Fig 1. Expression data was log2 transformed to facilitate comparisons across race groups.
Immunohistochemistry
Surgical FFPE sections of tumor samples that were processed at UPHS (NAMSenegal n = 52, EAM n = 100, AAM n = 104) were selected for IHC staining. Fig 1 provides sample details, used in IHC staining. H-score was calculated based on both intensity and percentage of immunoreactivity of the staining cells for selected biomarkers of interest including ERG, PTEN, MYC, Racemase (AMACR), AR, EZH2, Ki67, PSMA, and P53. All immunohistochemical stains were performed using antigen retrieval reagents from Leica, on the Leica bond 3 auto stainer. The details of each antibody are tabulated and presented in supplementary text. The heat induced epitope retrieval (HIER) ER1/20 is equivalent to citrate buffer at pH6 and ER2/20 is equivalent EDTA buffer at pH9.
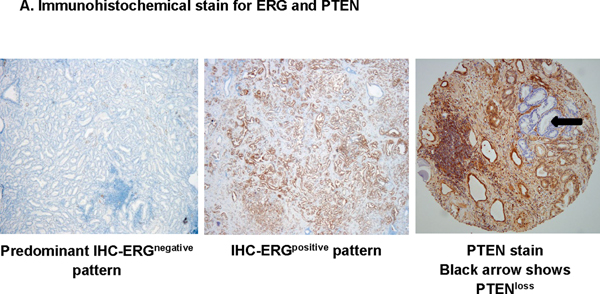
IHC scoring was based on the intensity of cytoplasmic staining compared to the surrounding stromal area and was semi-quantitatively scored on a scale of 0 to 3 by the pathologist. ERG-PTEN subtypes were derived based on the intensity of the ERG and PTEN. Samples that did not show any ERG expression (Intensity score = 0) were classified as ERGnegative, whereas samples with mild, moderate to significant expression (Intensity score 1 – 3) were ERGpositive (Fig 2A). Samples that exhibited complete loss (cytoplasmic intensity = 0) of PTEN protein in 100% of tumor cells were categorized as PTENloss, while remaining samples were grouped as PTENintact. Final categorization includes ERGnegative/PTENloss, ERGpositive/PTENloss, ERGnegative/PTENintact and ERGpositive/PTENintact (Fig 2A).
Figure 2.
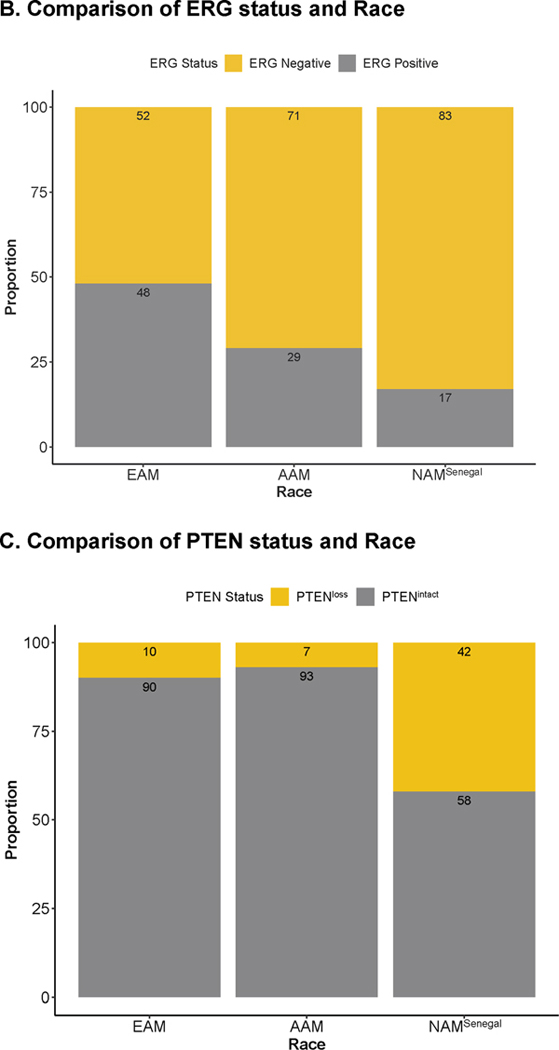
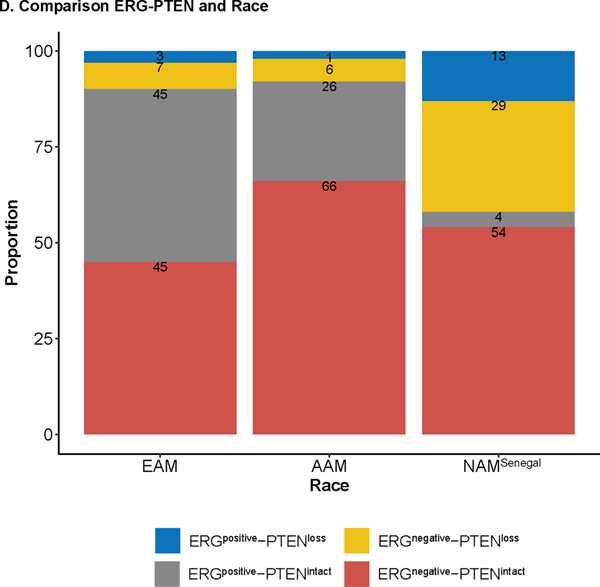
Immunohistochemical staining for the selected biomarkers using the samples processed at UPHS (n = 256).
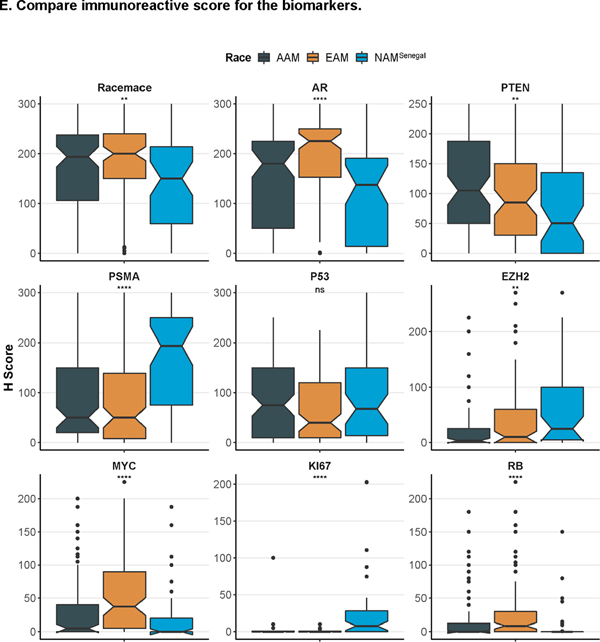
A. Immunohistochemical staining in prostate tumor sections for ERG and PTEN. B. Comparison of ERG status and Race; C. Comparison of PTEN status and Race. D. Comparison of ERG-PTEN and Race; E. Compare immunoreactive score for the biomarkers.
Additional data elements
Besides the genomic expression and IHC data, detailed information on age, Gleason score, pretreatment prostate-specific antigen (PSA), primary treatment, and use of androgen deprivation therapy was also available. Gleason scores were categorized as Grade Group 17.
Statistical Analysis
A detailed comparison of clinical characteristics of datasets was carried out using descriptive statistics. Among UPHS processed samples (NAMSenegal, EAM, and AAM), a contingency chi-square test was used to compare the association of IHC based subtypes and race groups. Additionally, Median H scores for the selected biomarkers were compared across race groups by Wilcoxon-rank test. Pathway exploration was performed within samples processed at MCC (NAMGhana and EAM, Fig 2) by comparing differentially expressed genes between NAMGhana and EAM using the Wilcoxon rank test. Benjamini-Hochberg method of false discovery rate (FDR) adjustment was used to account for multiple comparisons. A comparative gene expression analysis was performed to identify biologically distinct pathways within NAMGhana as compared to EAM. Log2 transformed expression across genes were compared and differentially expressed genes with a fold change of >0.1 and FDR < 0.05 were selected for subsequent Gene set enrichment analysis (GSEA). In GSEA we evaluated the overall enrichment of differentially expressed genes using HALLMARK and REACTOME pathways and identified the most common pathways in NAMGhana as compared to their European counterparts. We also created an androgen receptor (AR) signature score based on previously published canonical-AR target gene list 18,19. Intergene correlation with hierarchical clustering was used to identify highly correlated AR target genes clusters and Z score standardized cumulative expression of these genes were used to derive AR signature score. Median AR scores across race groups were compared using the Wilcoxon-rank test. All the analysis was conducted in R version 3.5.0.
Results
The entire study cohort included 408 PCa patients with enriched tumor samples of NAM from Ghana and Senegal. Samples processed at MCC underwent rigorous quality control (QC) metrics for RNA quality and integrity and were used in the genomic exploration of molecular pathways exploration in MSigDB (NAMGhana n = 106 and EAM n = 46), Fig 1. Disease and treatment characteristics of MCC cases included in gene expression analysis are shown in Table 1. Overall, a higher proportion of NAMGhana men were diagnosed at an older age (p = 0.03), had higher Grade Groups (p = 0.002) and pretreatment PSA (p < 0.0001), as compared to their US counterparts Table 1. Samples processed at UPHS were used in IHC based immunostaining analysis to compare selective biomarkers and to define the distribution of ERG-PTEN subtypes across race groups (NAMSenegal n = 52, EAM n = 100, and AAM n = 104), Table 1. Significant heterogeneity was observed in the UPHS cohort with lower percentage of AAM having high Gleason score (p = 0.04), whereas other correlated were similar between race groups.
Table 1.
Dataset characteristics
| MCC processed Samples | UPHS processed samples | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Characteristics | EAM (n=46) | NAMGhana (n=106) | P | AAM (n=104) | EAM (n=100) | NAMSenegal (n=52) | P * |
|
| |||||||
| Age Category | |||||||
| ≤60 | 13 (28.3%) | 12 (11.3%) | 0.03 | 57 (54.8%) | 50 (50.0%) | - | 0.7 |
| >60 – 70 | 23 (50.0%) | 46 (43.4%) | 3 (2.9%) | 2 (2.0%) | - | ||
| >70 | 10 (21.7%) | 35 (33.0%) | 44 (42.3%) | 47 (47.0%) | - | ||
| NA | - | 13 (12.3%) | 0 (0%) | 1 (1.0%) | 52 (100%) | ||
| Age | |||||||
| Median (IQR) | 64 (59 – 69) | 68 (64 – 73) | 0.02 | 58.5 (53.7 – 64) | 60.0 (56 – 65.5) | - | 0.07 |
| Grade Group (GG) a * | |||||||
| GG1–GG2 | 33 (71.7%) | 48 (45.3%) | 0.002 | 86 (82.7%) | 70 (70.0%) | - | 0.04 |
| GG3–GG5 | 13 (28.3%) | 58 (54.7%) | 17 (16.3%) | 29 (29.0%) | - | ||
| NA | - | - | 1 (1.0%) | 1 (1.0%) | 52 (100%) | ||
| Clinical PSA | |||||||
| Median (IQR) | 4.66 (3.5 – 6.8) | 33.2 (15.3 – 83.8) | < 0.0001 | 6.35 (4.80 – 9.75) | 5.70 (4.2 – 8.7) | - | 0.1 |
| Clinical PSA Category | |||||||
| 0 – 10 | 41 (89.1%) | 10 (9.4%) | < 0.0001 | 10 (9.6%) | 3 (3.0%) | - | 0.1 |
| >10 – 20 | 4 (8.7%) | 24 (22.6%) | 77 (74.0%) | 80 (80.0%) | - | ||
| >20 | 1 (2.2%) | 54 (50.9%) | 15 (14.4%) | 17 (17.0%) | - | ||
| NA | - | 18 (17.0%) | 2 (1.9%) | 0 (0%) | 52 (100%) | ||
| Tissue source | |||||||
| Biopsy | 43 (93.5%) | 92 (86.8%) | 0.3 | - | - | 21 (61.5%) | - |
| RP | 3 (6.5%) | 14 (13.2%) | 104 (100%) | 100 (100%) | 20 (38.5%) | ||
Abbreviations: AAM, African American men; EAM, European American men; NAM, Native African men; IQR, Inter quartile range; PSA, Prostate Specific Antigen; RP, Radical Prostatectomy.
Gleason Score were centrally reviewed by institutional pathologist
Grade Group (GG) 1–2 are defined as those with Gleason score 6(3+3) & 7(3+4); GG 3–5 included Gleason score ≥7(4+3)
NAMSenegal sample do not have baseline and clinical information.
p-value in UPHS cohort is estimated for the comparison between AAM and EAM only.
NA were excluded in the p-value estimation.
Distribution of Biomarkers across race groups
IHC analysis on selected biomarkers processed at UPHS showed a consistent association between ERG status (a surrogate for the TMPRSS-ERG translocation) and race.83% of west African men (NAMSenegal) had tumors that lacked ERG translocation (ERGnegative) as compared to AAM (71%) and EAM (52%), Fig 2B. More importantly, loss of PTEN was more prevalent in NAMSenegal (42%) whereas PTEN status was largely intact in both AAM and EAM (Fig 2C). Furthermore, a higher proportion of NAMSenegal (29%) was found to be double negative (ERGnegative and PTENLoss) whereas the proportion of double negative subtype was relatively similar between AAM (6%) and EAM (7%) (Fig 2D). When compared based on H-score across biomarkers, NAMSenegal tumors had significantly higher immunoreactivity (H-score) for PSMA, and EZH2 and revealed a higher proliferative index as seen by Ki67 expression, on the other hand, immunoreactivity was lower for PTEN, MYC, AR, RB, and Racemase, (all p < 0.05) (Fig 2E). Table S2 provides association between race and IHC based biomarker after adjusting for the grade group. In the multivariable linear regression model adjusted for grade group and PSA, AR and RB were consistently associated with the race group (Table S2).
Comparative genomics to uncover pathways enrichment in NAM
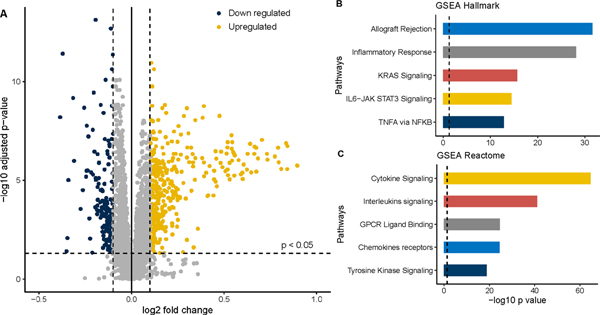
To focus the analysis on black men from West Africa with PCa, genomic expression analysis was compared between EAM and NAMGhana, using the samples that were processed at MCC. To ensure expression comparability across race groups, we compared the median expression of major housekeeping (HK) gene GAPDH and PPIA to show that the relative expression of HK genes was similar across race groups (Fig S1). Genomic expression of select biomarkers between race groups was compared within their respective panels i.e., OBP and PIP. From the OBP and PIP gene panels, a total of 559 genes were differentially expressed between EAM and NAM at FDR < 0.05 and fold change ≥ 0.1, including 377 upregulated and 182 downregulated genes (Fig 3A). In subsequent pathway enrichment analyses, upregulated genes in NAM (n = 377) were included in the GSEA using the molecular signature database (Midge). Within the HALLMARK pathways, allograft rejection, inflammatory response, KRAS signaling, TNFA signaling, IL6-JAK STAT3, complement, and interferon-gamma response were the highly upregulated pathways (Fig 3B). Similarly, within the REACTOME pathways, there were significant upregulation of cytokine signaling in the immune system, interleukin signaling, tyrosine receptor, FLT3 signaling, MAP kinase, IL-4 & IL-13, and GPCR ligand binding pathways (Fig 3C).
Figure 3.
Comparative genomics of NAM and EAM using samples processed at MCC (n = 152).
A. Volcano plot showing the up and downregulated genes in NAM (EAM is a reference); B. Gene set enrichment analysis (GSEA) using Hallmark pathways; C GSEA using Reactome pathway. All the pathways are upregulated in NAM as compared to EAM.
Differential Canonical-AR signaling with NAM
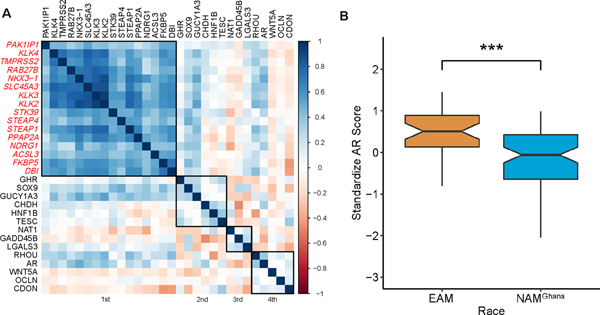
To assess the transcriptomic variability in androgen receptor (AR) activity across race groups we utilized a gene list of canonical AR target genes published earlier by Yamoah et al19 and Spratt et al20. Using the samples processed at MCC we performed intergene correlation and hierarchical clustering on mRNA expression of selected genes and identified a set of highly correlated AR target gene clusters (Fig 4A, genes within the first cluster). A composite mean expression score of these clustered genes - PAK1IP1, KLK4, TMPRSS2, RAB27B, NKX3−1, SLC45A3, KLK3, KLK2, STK39, STEAP4, STEAP1, PPAP2A, NDRG1, ACSL3, FKBP5, and DBI – were standardized to create an AR-activity score and compared across race groups. Like the findings from IHC-AR (Fig 2E), compared to EAM, NAMGhana had significantly lower AR-activity score (0.51 vs −0.08, p = < 0.0001), Fig 4B. In a Grade Group, pretreatment PSA and age adjusted multiple linear regression model, NAMGhana consistently showed a negative association with AR-activity score, Table S1.
Figure 4.
Androgen Receptor Signaling among samples processed at MCC (n = 152).
A. Intergene correlation between AR targeted genes. B. Comparison of the standardized AR scores between EAM and NAM.
Discussion
Enriched biomarkers in Native African Men suggest distinctive biology:
IHC based results presented herein suggest distinct biology in NAM characterized by the dysregulation of several biomarkers. TMPRSS2-ERG2 fusion is an early event in the development of prostate tumors and is regulated by AR receptor 13. While a large body of research has shown racial heterogeneity in TMPRSS2-ERG fusion, which is primarily absent (ERGnegative) in AAM 12,13, very little is known among native west African men 1. Earlier work by Zhou and colleagues showed a significantly lower prevalence of TMPRSS2-ERG in NAM, however, the study did not find any association between fusion status and high Gleason scores 1. Our results corroborate the findings from Zhou et al and showed that the proportion of ERGPOS tumors was indeed lowest in NAM, followed by AAM and EAM. Besides, a significantly higher proportion of NAM had PTENLoss, an important tumor suppressor implicated in tumor progression and adverse outcomes 21. While earlier work by Tosoian et al demonstrated that PTENLoss in AAM was indeed associated with aggressive outcomes, the extent to which it drives PCa progression in NAM remains largely unexplored 22. Interestingly, in a combined analysis, NAM had tumors with a higher proportion of double negative tumor subtypes (ERGnegative-PTENloss) compared with other race groups, which has been shown to drive lethal outcomes in PCa 21. Interestingly, NAM also had a higher expression of EZH2 (Enhancer of zeste homolog 2), a known transcriptional regulator involved in modulating the expression of several genes and tumor immune microenvironment through epigenetic machinery 23. Additionally, EZH2 expression is also associated with the metastatic progression of prostate tumors 24. Also, tumors from NAM exhibited dysregulation of several biomarkers including PSMA, RB1, and MYC which could have implications in PCa progression and aggressive outcomes 25–27.
Role of immune-biologic pathways in NAM
Enrichment analysis using differentially expressed genes that were upregulated in NAM revealed a unique repertoire of both classical HALLMARK as well as major immune signaling pathways. Most importantly, there is a significant enrichment of pro-inflammatory pathways including cytokine signaling, interleukins, and interferon-gamma response in NAM. Additionally, the gene expression profile of NAM prostate tumors is also highly enriched for the receptor tyrosine kinase as well TNFA signaling via NFKβ pathways, both of which are associated with cancer progression 28,29. Although immune infiltrates within the tumor microenvironment (TME) are associated with improved outcomes in other cancers, emerging studies have shown a negative association of immune enrichment and disease progression in PCa 30. Our results demonstrate that prostate tumors from NAM have an active immune microenvironment, which is hypothesis-generating and may provide mechanistic insights on the observed disparity in prostate cancer etiology and response to therapy. Recent work by Sartor et al demonstrated that AAM who received sipuleucel-T cell had better survival outcomes than EAM 9. Although not functionally validated, it plausible that PCa in men who share African ancestry have enrichment of immune-related genes, and that these tumors may respond favorably to immunotherapies.
Racial differences in AR signaling
The role of AR signaling is well characterized in both treatment naïve and castration-resistant metastatic PCa (mCRPC) 11,18. AR-mediated transcriptional machinery has been linked to immune-oncologic signaling including DNA repair pathways which are known to be dysregulated among men of African origin 20. Additionally, AR-dependent transcriptomic expression of various genes, including TMPRSS2-ERG, has also been implicated in the distinct PCa molecular subtypes among AAM 19,31. Emerging research has identified divergent AR signaling and DNA repair as a molecular basis of increased radiosensitivity among AAM 18,20,32. While functional validation of this association is warranted, the expression level differences in AR signaling - as shown in our work – and their role in radiation response will continue to evolve. Consistently, our results suggest that not only is AR signaling dysregulated in NAM, AR-activity is also significantly lower among NAM 20. Furthermore, the association was not driven by the underlying clinical differences among race groups, instead, NAM were independently associated with lower AR-activity. Taken together, our results support the need to further validate the significance of biological differences in the management of PCa in the NAM, as their prostate tumors may respond differently to emerging targeted therapies.
Results presented herein are first to characterize tumor microenvironment in PCa tumors from NAM which shows that these tumors exhibit dysregulation of several important biomarkers enriched for proinflammatory cytokines. Additionally, our study shows that NAM has a lower AR-activity score which could have implications on response to radiotherapy. However, the results are hypothesis-generating and exploratory, and further studies in much larger datasets are needed to evaluate the translational relevance and clinical implications of our results in the emerging landscape of precision oncology in PCa. Understanding these molecular mechanisms in context to less screened NAM, who are likely to present with more advanced PCa and share ancestral similarities to AAM in the US, may provide a unique opportunity to unravel the biologic basis of both incidence and outcome level disparities in AAM. Limitations to this study include the relatively small sample size, inadequate tissue from predominantly biopsy-derived tumors, and the lack of surrogate endpoints within a primarily radiotherapy treated cohort. Furthermore, the dataset used in the analysis are from two different clinical centers and unmeasured confounding factors resulting from the respective institutions are possible. Also, the study samples were selected based on the availability of biopsy tumors and therefore sample selection criteria could introduce some bias in the results. Native African Men also constitute an inherently aggressive subset of PCa with high PSA and these differences can influence the observed molecular and histochemical distinctions between race groups in this analysis. Lastly, we were unable to retrospectively determine genomic ancestry for all the samples due to limited tissue availability from primarily biopsy specimens. However, given the fact that the focus of the study was comparing NAM living in West Africa compared with EAM, we anticipate that genomic admixture (which is usually ~10% in AAM) will be minimal among the NAM population and will not impact our results to any major extent. More importantly, the cases included in the analysis were all centrally reviewed and analyzed on a single platform using stringent QC metrics, thus we are confident that our result provides insight into the genomic diversity at the transcriptome level of Black men living in African compared with predominantly White men in the US. Considering that NAM endures the highest burden of PCa globally, continued research is warranted to better understand the molecular framework of PCa race disparities.
Supplementary Material
Figure S1. Expression of housekeeping GAPDH and PPIA genes between EAM and NAM.
Acknowledgments
Funding: This work was supported by grant awards from The Prostate Cancer Foundation, P20-CA233255 (to K.Y. and T.R.R.), and H Lee Moffitt Cancer Center and Research Institute to Dr. Kosj Yamoah. This work is also supported in part by the Molecular Genomics Core Facilities at the Moffitt Cancer Center through its NCI CCSG grant (P30-CA76292).
Footnotes
Conflict of Interest: Relevant conflict of interest are reported in disclosure forms
Data Sharing Statement: Part of the data used in this study comes from an institutional registry and therefore cannot be shared. Queries regarding the data access for the MADCaP samples can be directed to www.madcapnetwork.org.
References
- 1.Zhou CK, Young D, Yeboah ED, et al. TMPRSS2:ERG Gene Fusions in Prostate Cancer of West African Men and a Meta-Analysis of Racial Differences. American journal of epidemiology. 2017;186(12):1352–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA: a cancer journal for clinicians. 2019;69(3):211–233. [DOI] [PubMed] [Google Scholar]
- 4.Globocan. Incidence, Mortality and Prevalence by cancer site. The Global Cancer Observatory. Ghana Web site. https://gco.iarc.fr/today/data/factsheets/populations/288-ghana-fact-sheets.pdf. Published 2018. Accessed2020. [Google Scholar]
- 5.Asamoah FA, Yarney J, Awasthi S, et al. Definitive Radiation Treatment Patterns and Outcomes for Low and Intermediate Risk Prostate Cancer Patients: A Cross-Continental Comparative Study. Am J Clin Oncol. 2019;42(12):937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baharian S, Barakatt M, Gignoux CR, et al. The Great Migration and African-American Genomic Diversity. PLoS genetics. 2016;12(5):e1006059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefflova K, Dulik MC, Barnholtz-Sloan JS, Pai AA, Walker AH, Rebbeck TR. Dissecting the within-Africa ancestry of populations of African descent in the Americas. PloS one. 2011;6(1):e14495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dess RT, Hartman HE, Mahal BA, et al. Association of Black Race With Prostate Cancer–Specific and Other-Cause Mortality. JAMA Oncology. 2019;5(7):975–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartor O, Armstrong AJ, Ahaghotu C, et al. Survival of African-American and Caucasian men after sipuleucel-T immunotherapy: outcomes from the PROCEED registry. Prostate cancer and prostatic diseases. 2020;23(3):517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halabi S, Dutta S, Tangen CM, et al. Overall Survival of Black and White Men With Metastatic Castration-Resistant Prostate Cancer Treated With Docetaxel. Journal of Clinical Oncology. 2019;37(5):403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahal BA, Alshalalfa M, Kensler KH, et al. Racial Differences in Genomic Profiling of Prostate Cancer. New England Journal of Medicine. 2020;383(11):1083–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamoah K, Johnson MH, Choeurng V, et al. Novel Biomarker Signature That May Predict Aggressive Disease in African American Men With Prostate Cancer. J Clin Oncol. 2015;33(25):2789–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faisal FA, Sundi D, Tosoian JJ, et al. Racial Variations in Prostate Cancer Molecular Subtypes and Androgen Receptor Signaling Reflect Anatomic Tumor Location. European urology. 2016;70(1):14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karakas C, Wang C, Deng F, Huang H, Wang D, Lee P. Molecular mechanisms involving prostate cancer racial disparity. Am J Clin Exp Urol. 2017;5(3):34–48. [PMC free article] [PubMed] [Google Scholar]
- 15.van Dessel LF, van Riet J, Smits M, et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nature Communications. 2019;10(1):5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odiaka E, Lounsbury DW, Jalloh M, et al. Effective Project Management of a Pan-African Cancer Research Network: Men of African Descent and Carcinoma of the Prostate (MADCaP). Journal of global oncology. 2018;4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egevad L, Delahunt B, Srigley JR, Samaratunga H. International Society of Urological Pathology (ISUP) grading of prostate cancer - An ISUP consensus on contemporary grading. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2016;124(6):433–435. [DOI] [PubMed] [Google Scholar]
- 18.Spratt DE, Alshalalfa M, Fishbane N, et al. Transcriptomic Heterogeneity of Androgen Receptor Activity Defines a de novo low AR-Active Subclass in Treatment Naïve Primary Prostate Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25(22):6721–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berglund AE, Rounbehler RJ, Gerke T, et al. Distinct transcriptional repertoire of the androgen receptor in ETS fusion-negative prostate cancer. Prostate cancer and prostatic diseases. 2019;22(2):292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spratt D, Dess R, Hartman H, et al. Androgen receptor activity and radiotherapeutic sensitivity in African-American men with prostate cancer: a large scale gene expression analysis and meta-analysis of RTOG trials. International Journal of Radiation Oncology• Biology• Physics. 2018;102(3):S3. [Google Scholar]
- 21.Ahearn TU, Pettersson A, Ebot EM, et al. A Prospective Investigation of PTEN Loss and ERG Expression in Lethal Prostate Cancer. Journal of the National Cancer Institute. 2015;108(2):djv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tosoian JJ, Almutairi F, Morais CL, et al. Prevalence and Prognostic Significance of PTEN Loss in African-American and European-American Men Undergoing Radical Prostatectomy. European urology. 2017;71(5):697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. 2018;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, Scott H, Carlsson SV, et al. Increased EZH2 expression in prostate cancer is associated with metastatic recurrence following external beam radiotherapy. Prostate. 2019;79(10):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebello RJ, Pearson RB, Hannan RD, Furic L. Therapeutic Approaches Targeting MYC-Driven Prostate Cancer. Genes (Basel). 2017;8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouchelouche K, Choyke PL, Capala J. Prostate specific membrane antigen- a target for imaging and therapy with radionuclides. Discov Med. 2010;9(44):55–61. [PMC free article] [PubMed] [Google Scholar]
- 27.Thangavel C, Boopathi E, Liu Y, et al. RB Loss Promotes Prostate Cancer Metastasis. Cancer Res. 2017;77(4):982–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Molecular cancer. 2018;17(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Molecular cancer. 2013;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao SG, Lehrer J, Chang SL, et al. The Immune Landscape of Prostate Cancer and Nomination of PD-L2 as a Potential Therapeutic Target. Journal of the National Cancer Institute. 2019;111(3):301–310. [DOI] [PubMed] [Google Scholar]
- 31.Wang BD, Yang Q, Ceniccola K, et al. Androgen receptor-target genes in african american prostate cancer disparities. Prostate cancer. 2013;2013:763569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matejcic M, Patel Y, Lilyquist J, et al. Pathogenic Variants in Cancer Predisposition Genes and Prostate Cancer Risk in Men of African Ancestry. JCO precision oncology. 2020;4:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression of housekeeping GAPDH and PPIA genes between EAM and NAM.