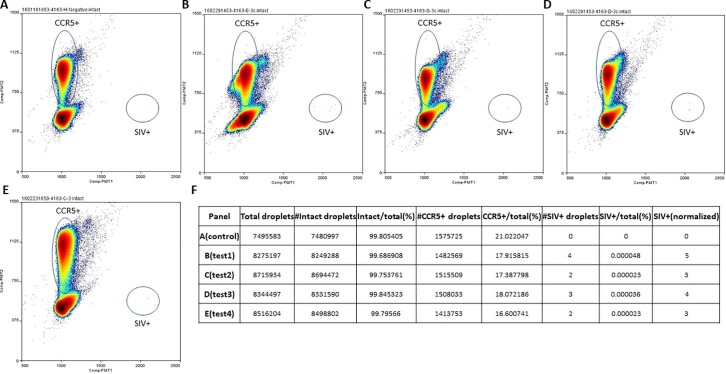
Fig. 3.
Low level SIV DNA signal detection on the Raindance platform using the SIV ddPCR assay. (A) A negative control sample which contains genomic DNA extracted from 1 million Rhesus macaque peripheral blood mononuclear cells (PBMC) from a naïve (i.e. uninfected) animal. (B-E) each corresponds to an average of 3 copies of SIV DNA standard spiked in genomic DNA extracted from 1 million Rhesus macaque PBMC from the same naïve animal as in (A). “CCR5+” and “SIV+” indicate CCR5 positive and SIV positive droplets, respectively. (F) Quantitation and statistics data corresponding to (A-E). The normalized SIV + values were calculated based on duplex Poisson adjustment as described in [20]. The average SIV + value for (B-E) was 4, and the coefficient of variation (CV) was 25.5%. Note that at the 3-copy target input level, the exact copies of the target templates that are present in the reactions follow a Poisson distribution, due to the stochastic limitations inherent in target sequence distribution in the volume of sample aliquots taken for testing.