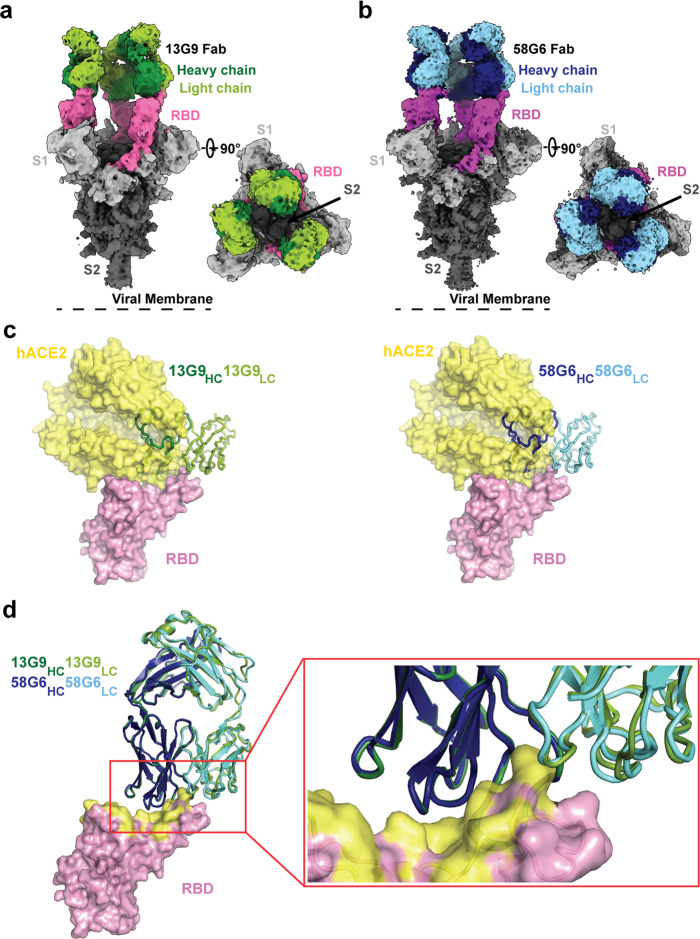
Fig. 4. Cryo-EM structures of 13G9 and 58G6 Fabs binding to open S trimer.
a, b Cryo-EM densities for 13G9 Fab (antigen binding fragment)-S (a; 3.9 Å) and 58G6 Fab-S (b; 3.6 Å) complexes, revealing binding of 13G9 or 58G6 to RBDs in the all ‘up’ state. c Superimposition of RBD-hACE2 [Protein Data Bank (PDB) ID 6LZG][https://www.rcsb.org/structure/6LZG] complex structure together with RBD-13G9 Fab (left) or RBD-58G6 Fab (right) variable domains, respectively. d Alignment of 13G9 and 58G6 Fabs on the same RBD. HC, heavy chain; LC, light chain. CDR, complementarity determining region.