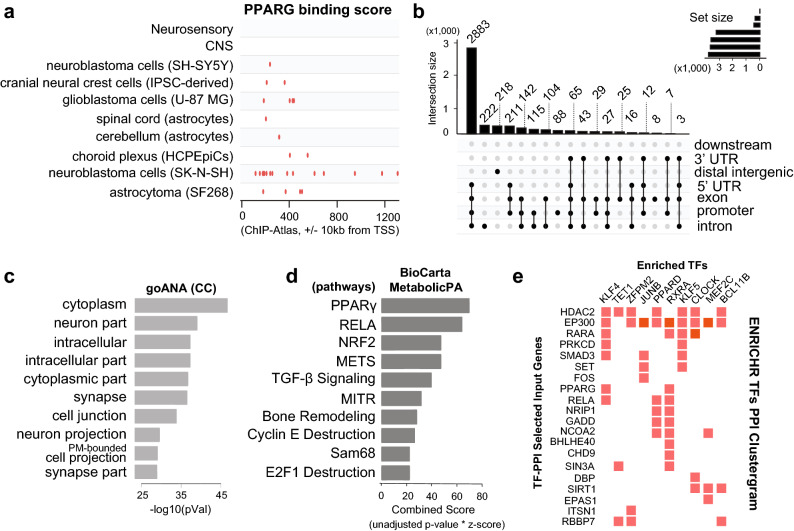
Figure 4 .
Characterization of genome-wide PARIS binding sites in SH-SY5Y neuroblastoma cells. (a) Human neuroblastoma cells provide a potential niche for PPARγ’s regulatory activities, justifying the choice of cell line for ChIP-seq experiment. (b) An upset plot19 showing the majority of significant PARIS ChIP-seq peaks overlap promoter region (− 3000, + 3000) of the genes in the human genome. (c) Enrichment of neuron-specific cellular components by PARIS taking the entire set of the peak-annotated genes as input. (d) Enriched metabolic pathways identified by BioCarta MetabolicPA module in EnrichR27 taking as input the significant peak-annotated genes that are also ranked by fold enrichment and that have a ChIP-seq peak overlapping the promoter region. Top 3 pathways include PPARγ and NRF2 pathways. (e) Enriched TFs identified by Transcription Factor PPI module in EnrichR27 taking the same genes in (d) as input. The most enriched TFs include PPARẟ and RXRA, the binding partner of PPARγ.