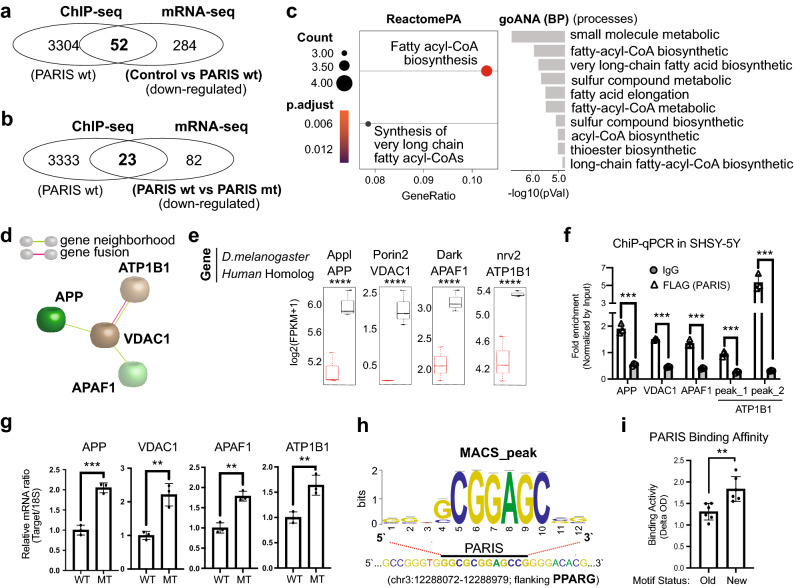
Figure 5 .
PPARγ orchestrates PARIS-driven transcriptomic changes. (a) A Venn diagram19 showing the shared genes between human orthologs of the DEGs identified as downregulated from “TRAP control versus PARIS wild type” comparison (TRAP-seq) and the entire set of the peak-annotated genes (ChIP-seq). (b) A Venn diagram19 showing the shared genes between human orthologs of the DEGs identified as downregulated from “PARIS C571A mutant versus PARIS wild type” comparison (TRAP-seq) and the entire set of the peak-annotated genes (ChIP-seq). (c) Ontology- and pathway-level enrichment results of the genes specified in (a). PPARγ-related GO terms are significantly enriched. (d-e) The STRING PPI network28 results of the shared genes specified in (b) and the expression profiles of the genes in this network. ***p ≤ 0.001, ****p ≤ 0.0001. The shared 23 DEGs successfully reproduced a part of the PPARγ network in Fig. 3a. FET p value threshold: 1e−03. Minimal Interaction Score: 0.400. Colored nodes: query genes (or proteins) and first shell of interactors. (f-g) ChIP-qPCR and RT-qPCR validation results of the target genes in (d). Both of the two distinct peaks associated with ATP1A1 gene were experimentally validated, together with the rest of the target genes. Statistical significances were analyzed with GraphPad Prism V7 software using Student’s unpaired t test. P values lower than 0.05 were considered significant. All qPCR-based expression data presented here were expressed as mean ± standard deviation values, for each condition studied. (h) PPARγ promoter region contains binding motif for PARIS. The k-mer significance = 123.19, the e-value = 6.5e−124, the number of genomic sites containing the motif = 4484. (i) The results of the colorimetric measurements that compare the binding affinities of PARIS to different motif sequences. PARIS has a significantly higher binding affinity to the new motif sequence identified within the scope of this study. **p ≤ 0.01.