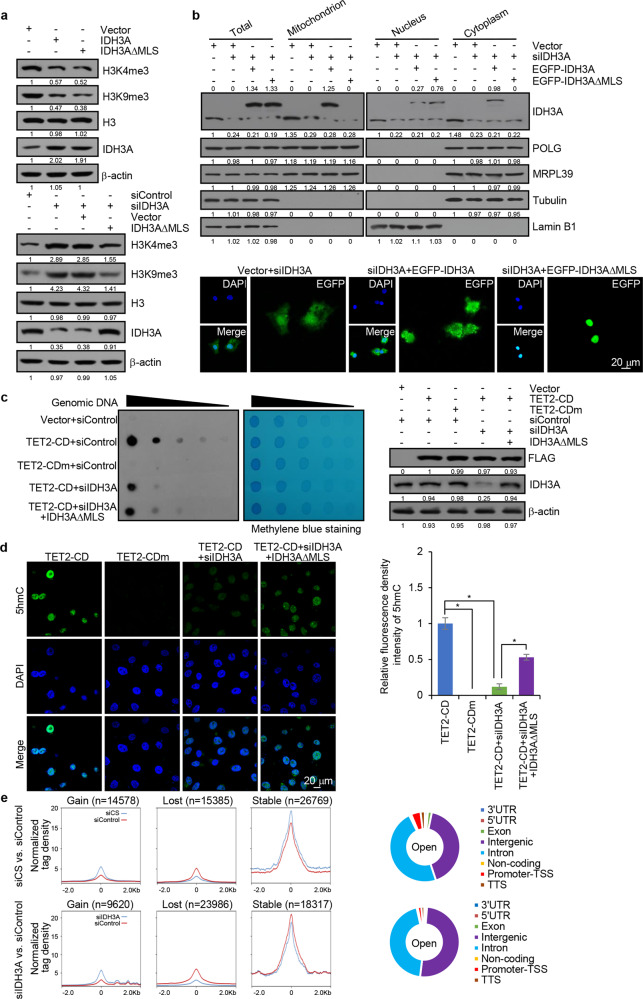
Fig. 5.
Nuclear IDH3A regulates histone/DNA methylation. a HepG2 cells were transfected with IDH3A, IDH3A∆MLS, and/or IDH3A siRNA for western blotting analysis with antibodies against the indicated histone marks or proteins. b HepG2 cells treated with IDH3A siRNA were transfected with EGFP-IDH3A or EGFP-IDH3A∆MLS for nuclei/cytoplasm/mitochondria isolation and western blotting analysis of IDH3A expression. The subcellular localization of IDH3A in these cells was analyzed by fluorescence microscopy. Bar, 20 μm. c HepG2 cells were co-transfected with FLAG-TET2-CD or FLAG-TET2-CDm and IDH3A siRNA and/or IDH3A∆MLS for dot blotting analysis with anti-5hmC. Spotted DNAs were stained with methylene blue to control the equal loading. The expression of the corresponding proteins was determined by western blotting. d HepG2 cells were co-transfected with FLAG-TET2-CD or FLAG-TET2-CDm and IDH3A siRNA and/or IDH3A∆MLS for immunofluorescent staining with anti-5hmC (green). DAPI staining was included to visualize the nucleus (blue). Bar, 20 μm. The relative fluorescent intensity of 5hmC was quantified by Image J software. Error bars represent mean ± SD for triplicate experiments (*p < 0.05). e ATAC-seq analysis in CS- or IDH3A-depleted HepG2 cells. The percentage of increased accessible sites in different genic regions is shown. UTR: 5′ and 3′ untranslated regions, TSS: transcription star sites, and TTS: transcription termination sites