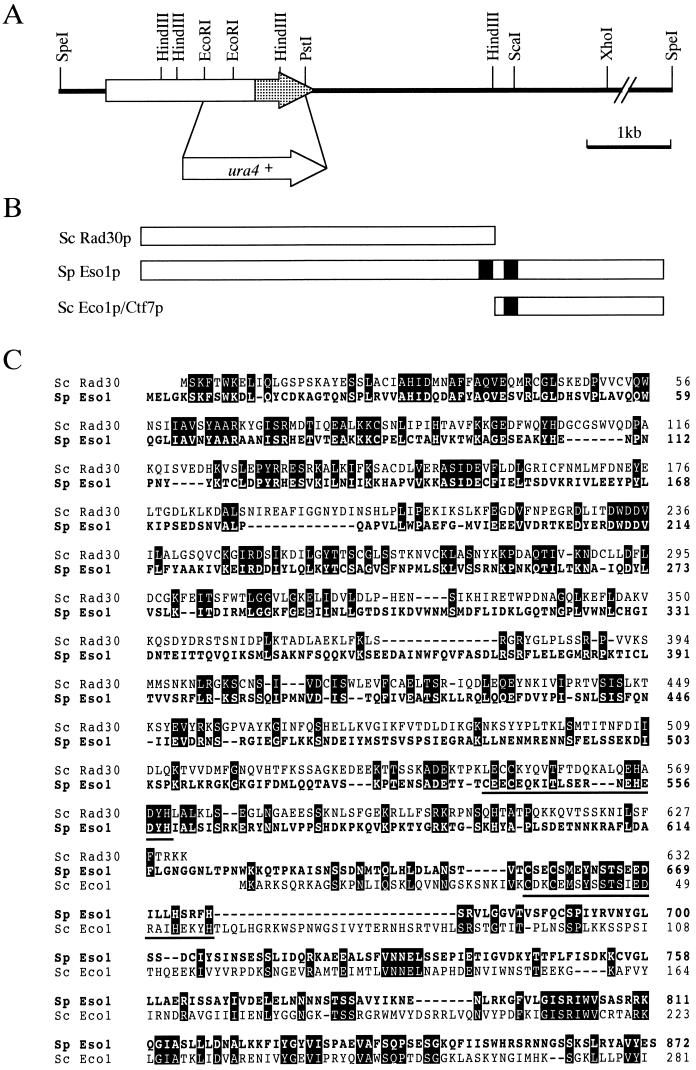
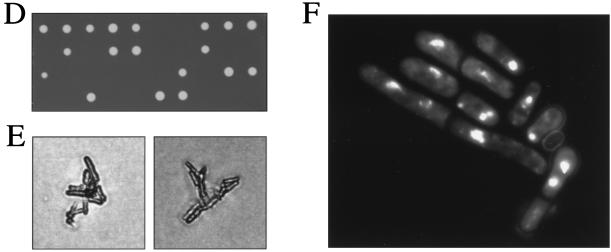
FIG. 5.
Isolation and characterization of the eso1+ gene. (A) Restriction map of the eso1+ gene. The eso1+ open reading frame is shown by an arrow, and the Eco1p/Ctf7p-homologous domain is shaded. The EcoRI-PstI region of eso1+ was replaced with a ura4+ gene cassette for generating an eso1 null mutant. (B) Schematic illustration of Eso1p of S. pombe and Rad30p and Eco1p/Ctf7p of S. cerevisiae. The zinc finger motifs are filled. (C) Amino acid homologies between Eso1p, Rad30p, and Eco1p/Ctf7p. The predicted amino acid sequence of Eso1p is shown in a single-letter code and aligned with Rad30p and Eco1p/Ctf7p. Identical amino acids are boxed. The zinc finger motifs are underlined. (D) Cells with deletions of eso1+ are lethal. Spores from eso1+/eso1::ura4+ diploid cells were tetrad dissected on YEA plates and incubated at 30°C for 4 days. (E) Terminal phenotype of Δeso1 cells. Cells that germinated from two independent Δeso1 spores on a YEA plate were photographed. (F) DAPI staining of germinating Δeso1 cells. Δeso1 spores (Ura+) derived from eso1+/eso1::ura4+ diploid cells were preferentially germinated in MM lacking uracil. Germinating cells were fixed with 70% ethanol, stained with DAPI, and photographed under the microscope.