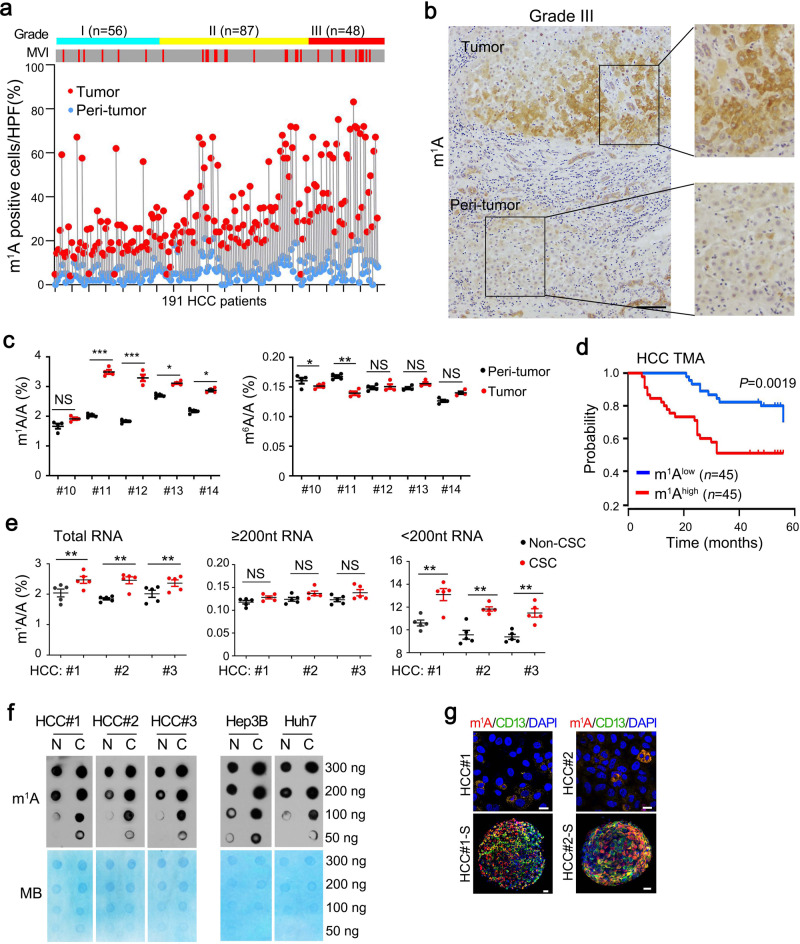
Fig. 1. m1A methylation in RNA are aberrantly elevated in HCC tumor tissues and liver CSCs.
a Overview of immunohistochemistry analysis of endogenous m1A levels in a panel of 191 clinically defined human HCC samples. Samples are arranged by grades and then sorted by microvascular invasion (MVI) (gray: “no”; red: “yes”). Paired samples are connected with gray lines. b Representative immunohistochemistry images of endogenous m1A levels (brown) in grade III HCC tumor tissue. Scale bar, 100 μm. n = 4 independent experiments. c LC-MS/MS quantification of m1A/A and m6A/A ratios in total RNA purified from HCC samples. Data are means ± SD. n = 4 biologically independent experiments. Exact P values from left to right: 0.052, 0.00082, 0.00043, 0.026, 0.011, 0.043, 0.0075, 0.13, 0.084, 0.055. d Kaplan–Meier plots of overall survival based on m1A levels in the tissue microarray assay (TMA) cohort. The median expression value is used as a cut-off. P values for Kaplan–Meier curves were calculated using a log-rank test. e LC-MS/MS quantification of m1A levels in total RNA, large RNA (≥200 nucleotide, nt, mostly composed of rRNA) and small RNA (<200 nt, mostly composed of tRNA) purified from HCC primary CSCs and non-CSCs, presented as percentage of unmodified A. Data are means ± SD. n = 5 biologically independent experiments. Exact P values from left to right: 0.0052, 0.0063, 0.0065, 0.067, 0.11, 0.075, 0.0017, 0.0089, and 0.0084. f Global m1A levels were detected in liver CSCs (C) and non-CSCs (N) from HCC primary tumors using dot blot assay. MB (methyl blue) was used as loading controls. n = 4 biologically independent samples. g Representative immunofluorescence staining of m1A and CD13 in HCC primary tumor cells and respective oncosphere cells (S). DAPI, 4,6-diamidino-2-phenylindole. Scale bar, 10 μm. n = 4 biologically independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, and NS, not significant (P > 0.05) by two-tailed Student’s t-test.