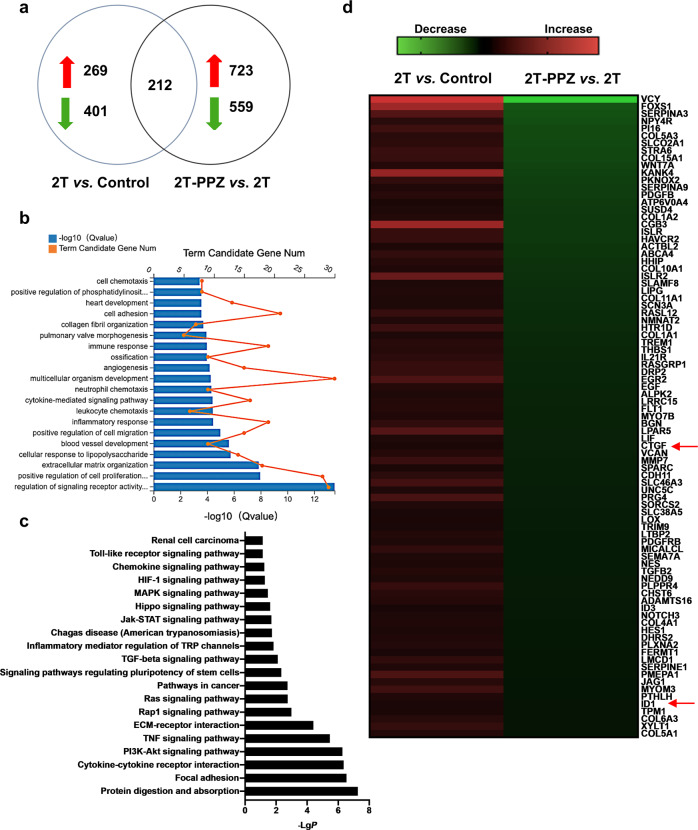
Fig. 4. Transcriptome analysis of LX-2 cells.
LX-2 cells were starved for 24 h and then treated with 2 ng· mL−1 TGFβ1 (2 T) and 200 µM PPZ for an additional 24 h. The total RNA was extracted for transcriptome analysis. a Number of genes significantly altered in the transcriptome analysis. b Gene ontology (GO) analysis (biological process) based on the classification of gene numbers, expression, and significance probability. c KEGG pathways were classified by DAVID functional annotation bioinformatics of differential genes in transcriptome analysis. d Expression profiling and heat map of the abovementioned grouping of 60 fibrogenesis-related genes.