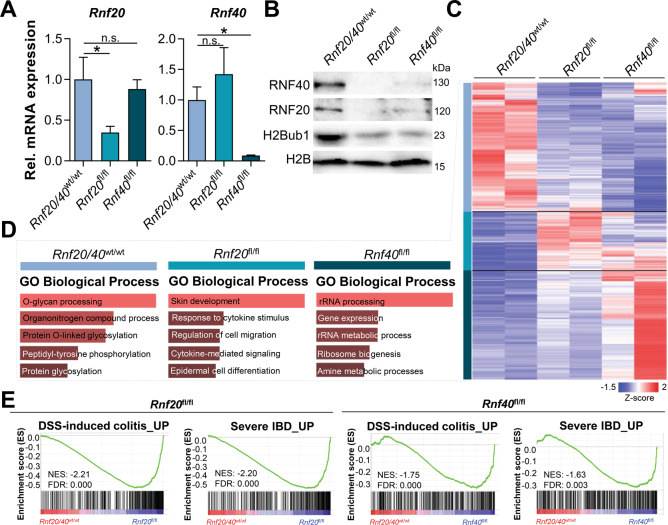
Fig. 3. The intestinal loss of either Rnf20 or Rnf40 drives IBD-associated transcriptome-wide changes.
A The loss of Rnf20 or Rnf40 in IECs was confirmed at the mRNA and (B) protein levels using qRT-PCR and western blot, respectively. C mRNA-seq was performed using wild type, Rnf20fl/fl and Rnf40fl/fl IECs (n = 2). Heatmap depicting the transcriptome-wide effects of Rnf20 and Rnf40 deletion by showing significantly (padj ≤ 0.05) upregulated (log2 FC ≥ 1, red) or downregulated (log2 FC ≤ −1, blue) genes. D Based on expression patterns, genes were divided into three clusters which were analyzed using gene ontology (Enrichr) [21, 22]. Genes enriched in wild type controls were associated with O-glycosylation while genes enriched in Rnf20 and Rnf40 knockout mice correlated with cytokine-mediated signaling and gene expression processes, respectively. E Differentially regulated genes upon Rnf20 and Rnf40 knockout in IECs were compared to previously published gene expression signatures [23, 24] using GSEA. Transcriptional changes associated with DSS-induced colitis in mice as well as severe IBD were enriched in Rnf20fl/fl and Rnf40fl/fl IECs. One-way ANOVA, mean ± SEM.