Fig. 1.
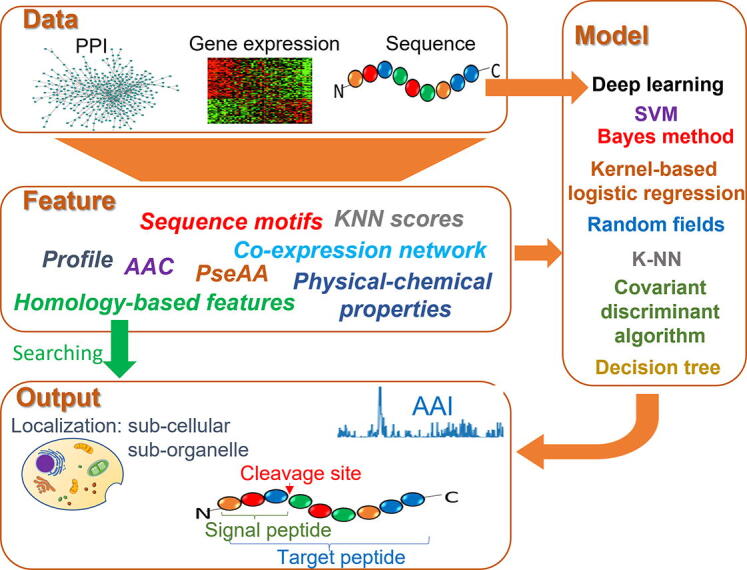
Relationships among the data, features, models, and prediction outputs in the computational prediction of protein localization. Sequence data can be converted into different features before feeding the data to a classifier model. Some classification models take raw data (e.g., one-hot-encoding of protein sequences for deep learning) as input, while others use engineered features. Localization prediction (at the sub-cellular and/or suborganellar level) is the most common output. Some methods also provide side product predictions such as target peptides, signal peptide cleavage sites, and mechanism interpretability at amino-acid-level resolution (AAI). Homology-based methods are special in the sense that they can make predictions directly based on homology-based features, such as the GO terms of homologous proteins.
