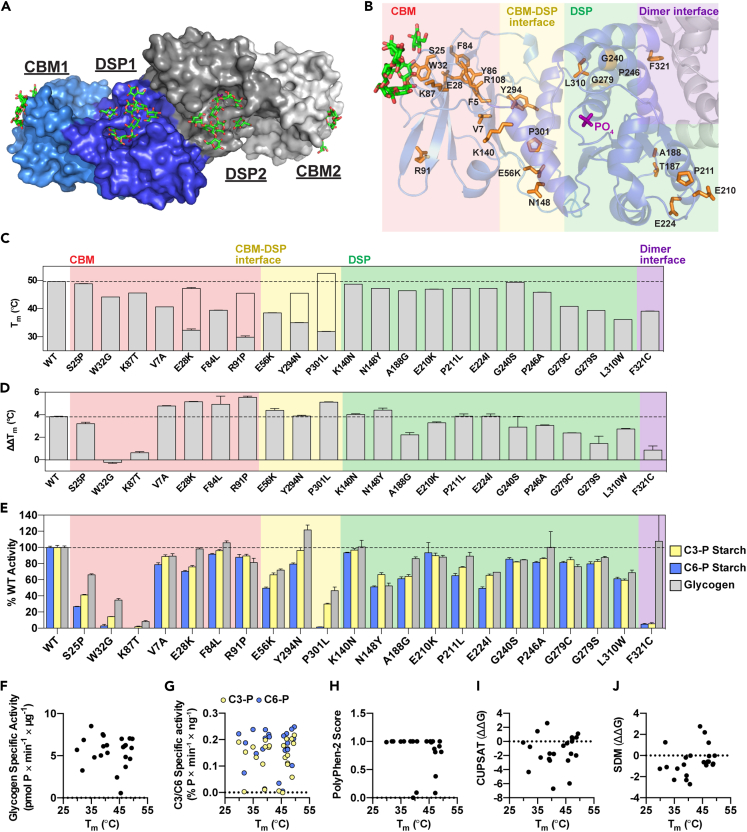
Figure 2.
Stability, carbohydrate binding and phosphatase activity of LD mutants
(A) Surface structure of laforin bound to maltohexaose, i.e. DP6 (PDB:4rkk) (Raththagala et al., 2015). One homodimer is shown with the CBM and DSP domain of one subunit shown in light and dark shades of blue, respectively. Maltohexaose molecules, shown in green, were bound to the CBM and DSP domain in each subunit of the dimer.
(B) Ribbon diagram of one subunit of the laforin dimer. Residues affected by missense mutations are shown in orange. Phosphate bound to the active site is shown in purple, and glucans bound to the CBM are shown in green. Mutations in the CBM are boxed in pink, those in the CBM-DSP domain interface in yellow, those in the DSP domain in green, and those in the dimer interface in purple.
(C) Mutant stability measured by melting temperature (Tm). For mutants displaying biphasic melting, the first (primary) peak is represented by the filled gray bar; the empty bar indicates the approximate Tm corresponding to the second peak.
(D) The difference in ΔTm displayed for each mutant in the presence of 10 mM DP7 compared to 10 mM DP24, indicated as ΔΔTm.
(E) Specific activity of laforin mutants with the indicated substrates. Activity of each mutant was normalized to WT activity. In (C), (D) and (E), all assays were performed in triplicate, graphs represent the average ± SD, and WT levels are indicated with a dashed line.
(F–J) Correlation scatterplots of Tm versus glycogen specific activity (r = −0.005929, p = 0.9786); C3-P and C6-P starch specific activity (r = 0.2141, p = 0.3266 for C3-P; r = 0.09289, p = 0.6734 for C6-P); PolyPhen-2 score (r = −0.4068, p = 0.0603), and CUPSAT (r = 0.2033, p = 0.3641) and SDM (r = 0.3972, p = 0.0672) predictions for ΔΔG (kcal/mol). Also see Figure S1.