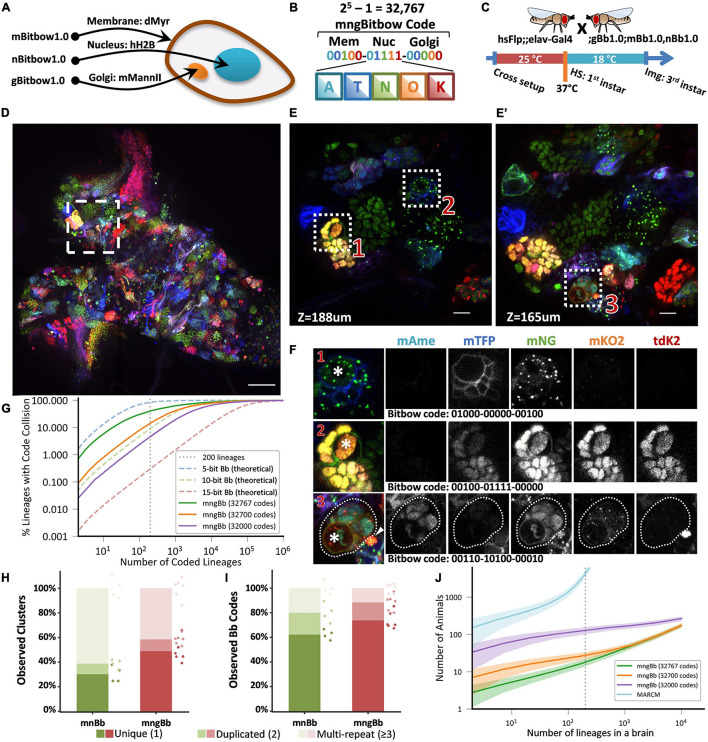
FIGURE 2.
Targeting Bitbow fluorescent proteins (FPs) to multiple subcellular compartments enables high-throughput lineage tracing without ambiguity. (A) The same Bitbow FPs are targeted to cell membrane, nucleus, or Golgi apparatus to generate spectrally-spatially resolvable Bitbow codes. (B) Up to 32,767 unique mngBitbow codes can be generated can be presented as 3 groups of 5-digit 0/1s in correspondence with the expression status of mAmetrine (A), mTFP1 (T), mNeonGreen (G), mKO2 (O) and tdKatushka2 (K). (C) Experimental setup of generating heat-shock induced mngBitbow1.0 labeling and imaging. (D) Maximum intensity projection of a 3rd instar mngBitbow1.0 brain. Scale bar, 50 μm. (E,E’) Two confocal image slices corresponding to two different z positions in the boxed region in panel (B). Scale bar, 10 μm. (F) 3 clusters marked in panels (E,E’) are assigned mngBitbow barcodes. Asterisks indicate the neuroblasts of each cluster. The arrowhead highlights an adjacent neuroblast labeled by a distinct mngBitbow code. (G) Simulation of Bitbow code collision in lineage mapping experiments. Dashed curve lines are simulations based on theoretical Bitbow code frequencies. Solid curve lines are simulations based on observed Bitbow code frequencies. Vertical dotted line corresponds to mapping all of the 200 lineages in a single adult Drosophila central brain. (H) Percentages of cell clusters that are uniquely labeled, or 2 of them, or ≥3 of them are labeled by the same mnBitbow (286 clusters, 4 brains) or mngBitbow (577 clusters, 6 brains) codes in each brain. Means and all data points are shown. Each dot represents quantification from one brain, and dots are colored in the same way as the stacked bar graphs. (I) Percentages of mnBitbow (N = 80, 4 brains) or mngBitbow (N = 240, 6 brains) codes that are expressed in 1, or 2, or ≥3 clusters in each brain. Means and all data points are shown. Each dot represents quantification from one brain, and dots are colored in the same way as the stacked bar graphs. (J) Monte Carlo simulations estimate the number of animals that are needed (y-axis) to sample all lineages at least once in animal brains that have given numbers of lineages (x-axis). Solid lines, means. Shaded lines, SD.