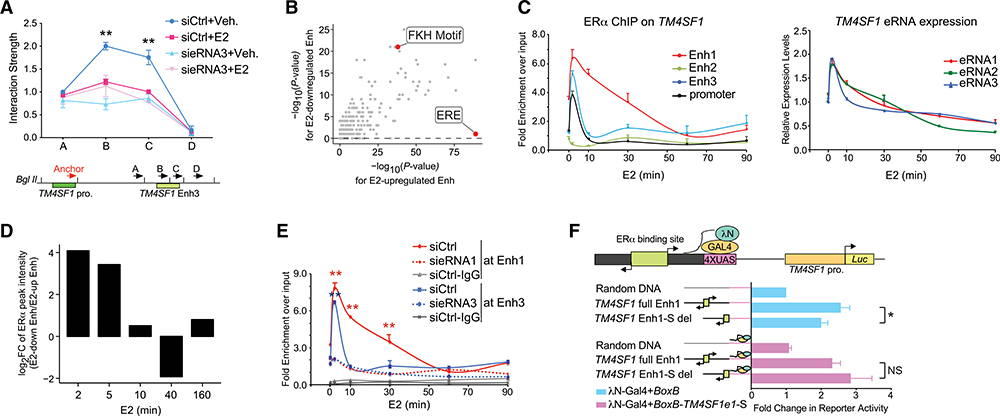
Figure 3. E2-Repressed eRNAs Assist in Recruiting ERα to Specific Enhancers.
(A) 3C analysis of the interaction between the TM4SF1 promoter (TM4SF1 pro.) and Enh3 upon knock down of eRNA3 in MCF-7 receiving vehicle (Veh.) or 100 nM E2 for 3 h. Bottom: illustrative diagram of the anchor region and selected docking sites (A–D).
(B) Scatterplot showing the significance of transcription factor binding motifs in E2-downregulated (y axis) and E2-upregulated (x axis), ERα-bound enhancers. Forkhead (FKH) motif and estrogen response element (ERE) were marked.
(C) ERα binding at indicated cis-regulatory elements near TM4SF1 gene (left) and expression of TM4SF1 eRNAs (right) in MCF-7 cells treated with 100 nM E2 for indicated lengths of time.
(D) FC of ERα ChIP-seq peak intensities at each indicated time point after E2 stimulation at E2-downregulated enhancers relative to those at E2-upregulated enhancers.
(E) E2-induced recruitment of ERα to Enh1 and 3 of TM4SF1 after knocking down indicated eRNAs in MCF-7 with 100-nM E2 treatment for various lengths of time. ChIP with normal IgG antibody in cells transfected with nontargeting siRNA (siCtrl) was included as a negative control.
(F) TM4SF1 native promoter (TM4SF1 pro.)-driven luciferase activity in the absence (blue bars) or presence (light pink bars) of BoxB-fused sense-strand eRNA1 of TM4SF1 (BoxB-TM4SF1e1-S) in MCF-7 cells treated with 100 nM E2 for 5 min. Top: schematic diagram of RNA-tethering luciferase assay.
Data in (A), (C), (E), and (F) are presented as mean ± SD with three biological and technical replicates. p values in (A), (E), and (F) were calculated using the t test and in (B) were obtained using the hypergeometric distribution in the HOMER program (Heinz et al., 2010). *p < 0.05; **p < 0.001. Also see Figure S4.