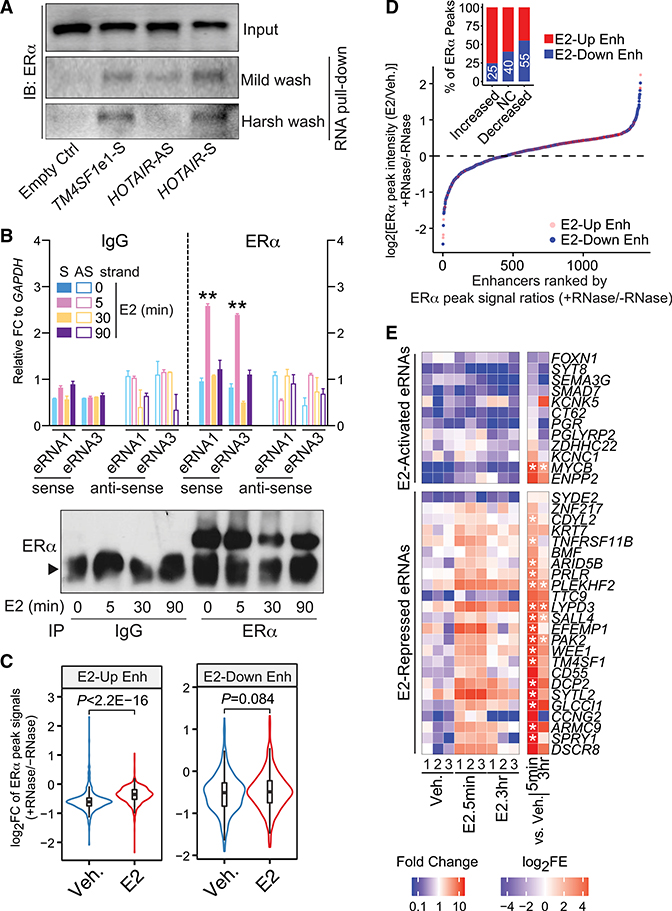
Figure 4. E2-Repressed eRNAs Directly Bind to ERα and Facilitate ERα Binding at E2-Downregulated Enhancers.
(A) In vitro RNA pull-down assay by incubating the in-vitro-transcribed, biotin-labeled TM4SF1 eRNA1 with the nuclear extract from MCF-7 cells treated with 100 nM E2 for 3 h. Empty Ctrl, streptavidin beads with no RNAs; TM4SF1e1-S, sense-strand eRNA1 of TM4SF1; HOTAIR-AS, antisense-strand HOTAIR as negative control; HOTAIR-S, sense-strand HOTAIR as positive control.
(B) RIP of ERα in MCF-7 cells treated with 100 nM E2 for indicated duration of time. Normal IgG antibody was included as a negative control, and data were presented as the FC to GAPDH mRNA levels. S, sense-strand; AS, antisense-strand. Bottom: western blot showing immunoprecipitation (IP) efficiency. Arrowhead, IgG heavy chain.
(C) Violin plot comparing the log2FC of ERα peak intensities before (−) and after (+) RNase addition under vehicle (Veh.) or E2 (100 nM for 30 min) treatment condition at E2-upregulated (E2-Up Enh) and E2-downregulated (E2-Down Enh) enhancers.
(D) Rank of ERα-bound, E2-upregulated (red dots), and E2-downregulated (blue dots) enhancers based on the ratios of E2-induced ERα chromatin binding intensities with RNase treatment (+RNase) to those without RNase treatment (−RNase). The E2-induced ERα chromatin binding was evaluated by comparing the ERα ChIP-seq peak signals between E2- and vehicle-treatment conditions (E2/Veh.). Insert: percentages of E2-downregulated (blue bars) and E2-upregulated enhancers (red bars) in each group of ERα ChIP-seq peak signals (E2/Veh.): increased, log2[ERα peak intensities (E2/Veh.) + RNase/ERα peak intensities (E2/Veh.) − RNase ≥ log2(1.2); no change (NC), −log2(1.2) ≤ log2[ratios] ≤ log2(1.2); or decreased, log2[ratios] ≤ −log2(1.2) after adding RNase compared to no RNase treatment. Numbers in white, the proportions of E2-downregulated enhancers in the aforementioned groups of ERα peaks.
(E) Heatmap showing eRNA enrichment in the ERα PAR-CLIP immunoprecipitates from MCF-7 cells that were treated with vehicle (Veh.) or 100 nM E2 for 5 min (E2.5min) or 3 h (E2.3hr). Data from three biological replicates (1, 2, and 3) were presented. FC, levels of specified eRNAs in the ERα immunoprecipitates upon E2 treatment compared to the vehicle condition; log2(fold enrichment) (FE), FE of indicated eRNAs in the ERα immunoprecipitates over the input RNAs. The final results in (B) and (E) were obtained from three biological and technical replicates; the data in (B) are presented as mean ± SD; P values in (B), (C), and (E) were determined using t test. *p < 0.05; **p < 0.001. Also see Figure S5.