Figure 4. Mechanisms resulting in biofilm dispersal.
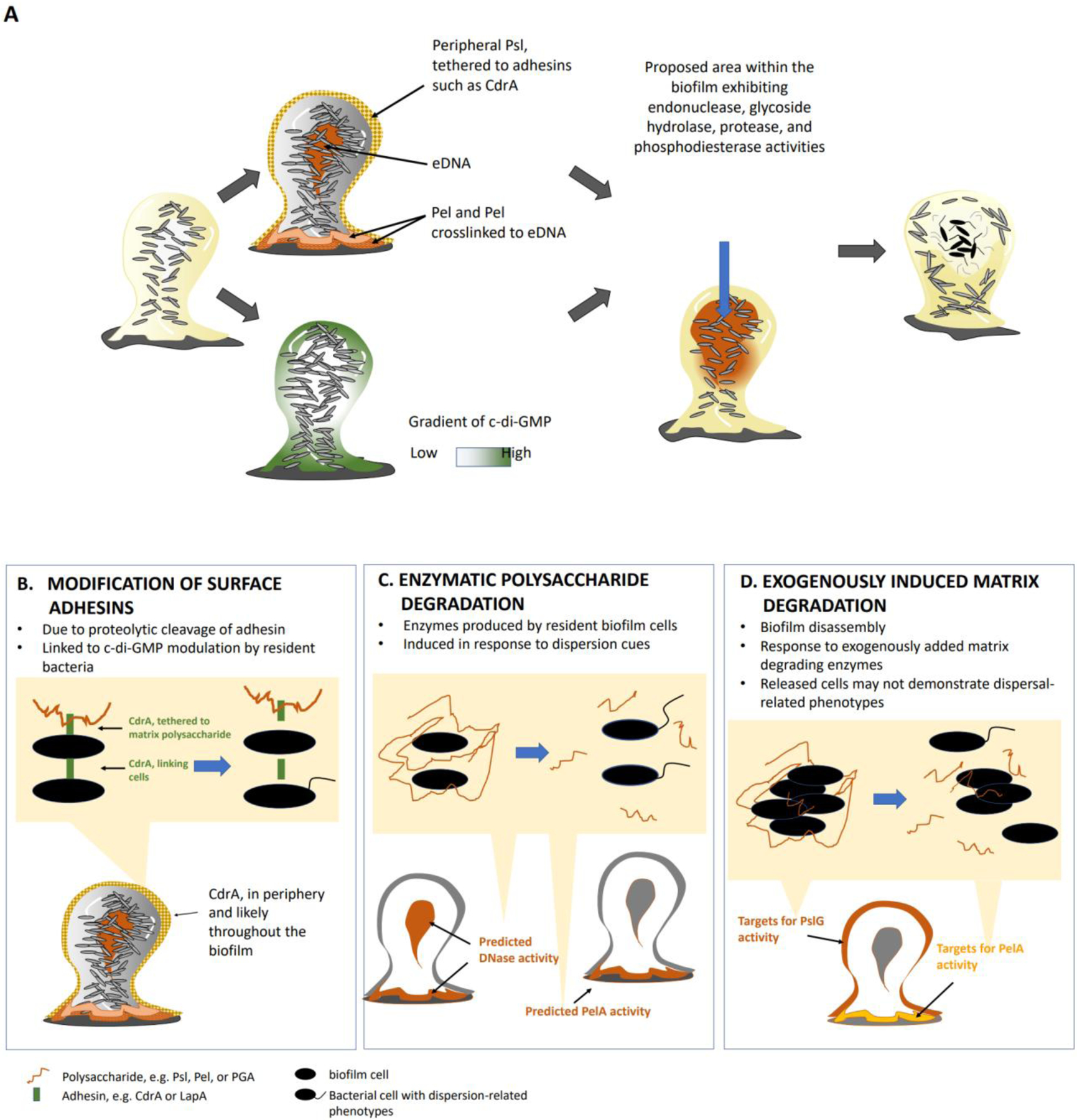
(a) Model of spatial localization of biofilm matrix components and bis-(3’-5’)-cyclic dimeric guanosine monophosphate (c-di-GMP), and proposed biofilm areas with active matrix- and c-di-GMP-degrading activities, that lead to biofilm dispersion. In Pseudomonas aeruginosa PAO1, the matrix polysaccharide Psl is localized at the periphery and the base of biofilms. Psl interacts with the adhesin protein CdrA and forms a protective but non-rigid structure in between cells and around the biofilm structure. The Pel polysaccharide is primarily located at the base of the biofilm and crosslinked to extracellular DNA (eDNA). However, eDNA is not limited to the biofilm base but has also been detected in the biofilm interior. Mature biofilms are characterized by low c-di-GMP levels in the interior of the biofilm whereas c-di-GMP levels in immature or less structured biofilms is more uniform. Considering the link between dispersion, low c-di-GMP levels, and matrix degradation, the biofilm periphery and biofilm interior are the most likely to be the locations within the biofilm that experiecne increased matrix- and c-di-GMP-degrading activities. Moreover, these locations coincide with observed dispersion events such as void formation and erosion of the biofilm structure. b) Surface adhesins such as CdrA or LapA are cleaved to untether the polysaccharide matrix and break cell-cell interactions. (c) Matrix components are enzymatically degraded (by intrinsic DNases or the hydrolase PelA) in response to native or environmental dispersion cues (nitric oxide, nutrients or cis-2-decenoic acid). d) Biofilm disassembly can be induced by exogenously added matrix-degrading enzymes. As enzymes are exogenously added, matrix degradation is expected to primarily target the periphery of the biofilm structure (see Box 1).
