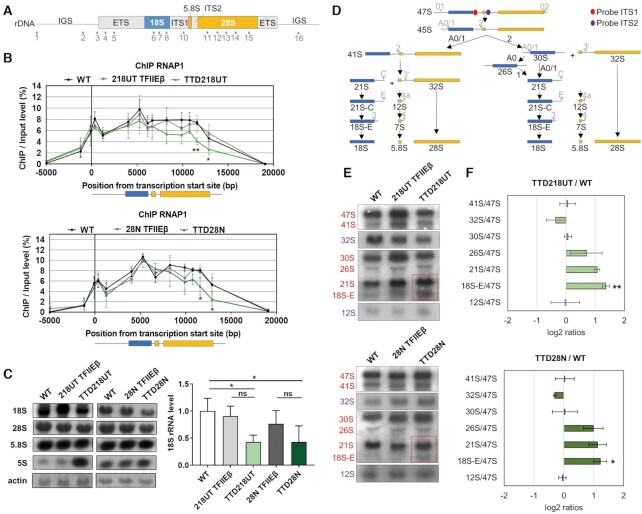
Figure 3.
TFIIE influences RNA polymerase I gene occupancy and rRNA processing. (A) Schema of the human rDNA unit. The non-coding region ETS, ITS1 and ITS2 (gray) and coding regions 18S (blue), 5.8S and 28S (both in orange) are illustrated. Primers used for ChIP analysis are indicated by arrowheads and numbers. Exact primer sequences are given in Table S2. (B) Quantitative qPCR ChIP analysis of RNAP1 in WT, reconstituted and TTD cells indicates reduced binding of RNAP1 to the end of the 28S rRNA coding rDNA (region 14 and 15) in TTD cells. (C) Left: Northern Blot analysis of mature 18S, 28S, 5.8S and 5S in WT, reconstituted and TTD cells. Right: Quantification of mature rRNA relative to actin shows decreased mature 18S level in TTD cells compared to WT cells. Quantification of 47S pre-rRNA, 28S, 5.8S and 5S mature rRNA are given in the Supplement S5B. (D) Schema of the rRNA processing pathway in human cells; adapted from (45). ITS1 and ITS2 probes positions are marked in red and purple, respectively. (E) Northern Blot of WT, reconstituted and TTD cells show accumulation of 21S and 18S-E levels in TTD cells. Red and purple color code indicates rRNA species detected by ITS1 and ITS2 probe, respectively. Full images of the probed membrane are given in the Supplement S5B. (F) Analysis of the northern blots in (E) are displayed as Ratio Analysis of Multiple Precursors (RAMP) profiles;(24). Data are represented as mean ± SD of at least three independent experiments. ns P > 0.05, P * P ≤ 0.05, ** P ≤ 0.01.