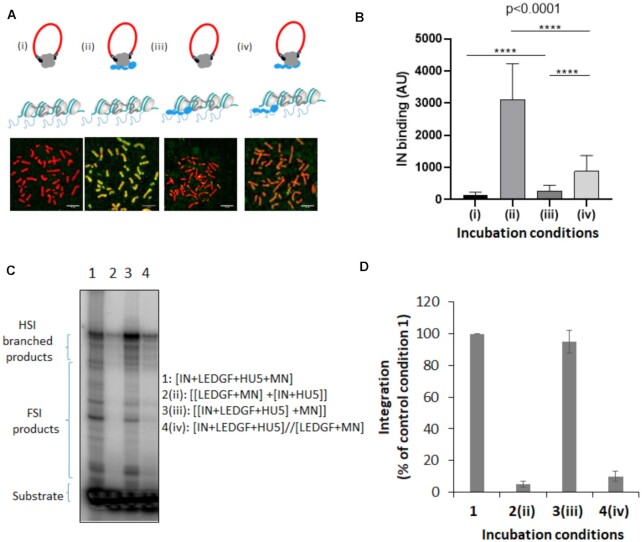
Figure 6.
Time of addition analysis of the LEDGF/p75-mediated chromatin tethering of HIV-1 IN. HIV-1 IN was incubated with chromosome spreads in the presence or in the absence of LEDGF/p75 following different time of addition conditions reported in (A): (i) IN alone, (ii) IN and LEDGF/p75 were preincubated before being added to the chromosome spreads, (iii) LEDGF/p75 was pre-incubated with chromosome spreads before adding IN and (iv) IN and LEDGF/p75 were preincubated together and added to chromosomes spreads previously precincubated with LEDGF/p75. The total amount of LEDGF was similar in all conditions. The intensity of total IN IF signal has been quantified in each condition using ImageJ software on several chromosomes and reported as IN binding. The data are reported in (B) as mean of the quantification from 12 chromosomes (here chromosome 3, 1135–1220 quantification points per condition) ± standard deviation (SD). Statistics were performed by student test and the calculated p is reported on the figure. Integration assay onto MN was performed in difference incubation conditions using recombinant IN with or without LEDGF/p75 as done in Figure 5. A representative gel is reported in (C) and quantification data obtained from at least three independent experiments are reported IN in the graphic (D) as means ± SD. Integration product structures are reported as half site integration (HSI) or full site integration (FSI). Condition 1: IN, LEDGF/p75, viral donor U5 DNA and target nucleosomal DNA were pre-incubated altogether, condition 2: [LEDGF/p75/target nucleosomal DNA] and [IN/viral U5 donor DNA] were preincubated separately before reaction, condition 3: IN, LEDGF/p75 and the viral donor U5 DNA were preincubated together before adding the target nucleosomal DNA, condition 4: [IN, LEDGF/p75, viral donor U5 DNA] and [LEDGF/p75, target nucleosomal DNA] were incubated separately before reaction.