Figure 1.
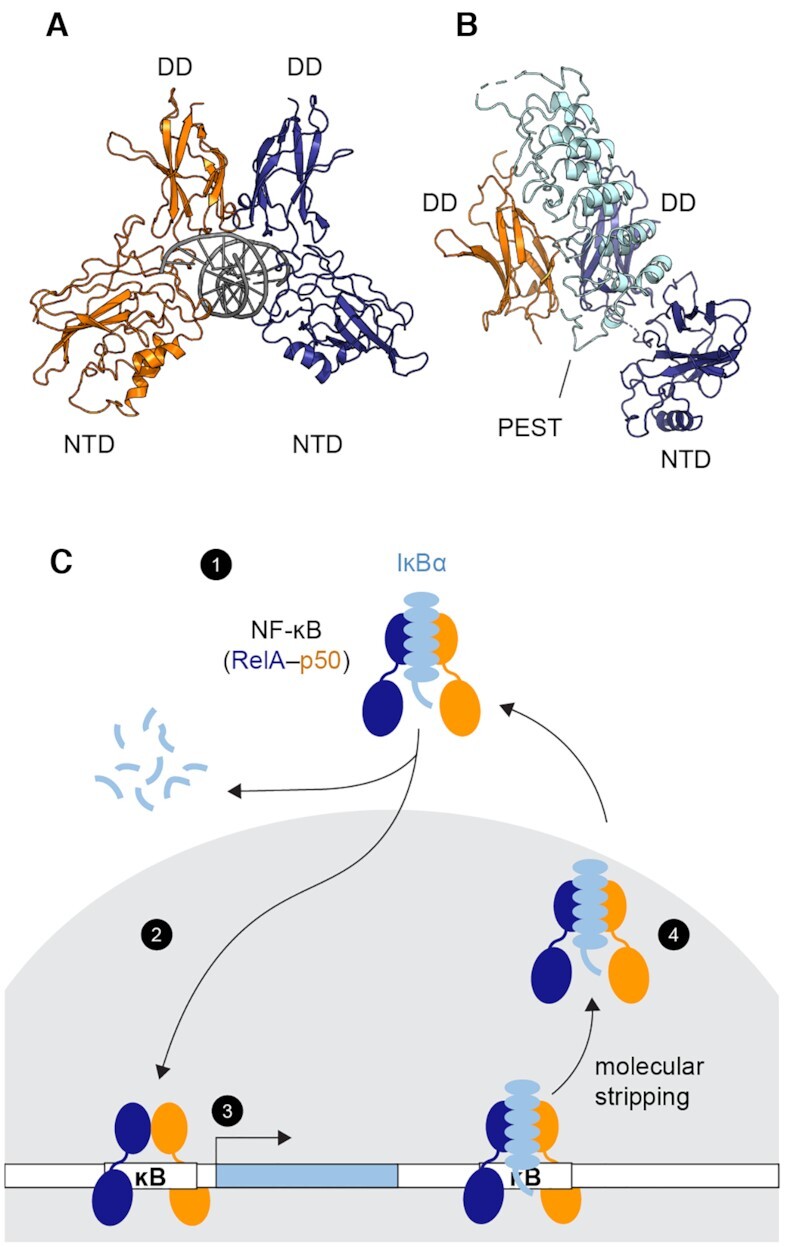
Transcriptional regulation by NF-κB. (A) Structure of the NF-κB RelA-p50 heterodimer bound to DNA (PDB: 1LE5, RelA in blue, p50 in orange and DNA in gray). Each monomer contains a dimerization domain (DD) and an N-terminal DNA binding domain (NTD) connected by a 10-amino-acid linker. The NF-κB dimer recognizes its cognate DNA through the two NTDs (B) Structure of the NF-κB bound to the inhibitor protein IκBα (PDB: 1IKN, IκBα in cyan). IκBα binds to the DDs of NF-κB. The disordered PEST sequence of IκBα inserts into the DNA-binding cavity. The NTD of p50 was truncated to achieve crystallization. (C) Schematic of NF-κB transcriptional regulation showing (1) NF-κB is held inactive in the cytoplasm by IκBα. (2) Upon stimulation, targeted degradation of IκBα allows free NF-κB to translocate to the nucleus (gray). (3) NF-κB binds DNA and activates genes including the one of IκBα, creating newly synthesized IκBα. (4) IκBα enters the nucleus, accelerates the dissociation of NF-κB from the DNA via molecular stripping, and returns NF-κB to the cytoplasm.
