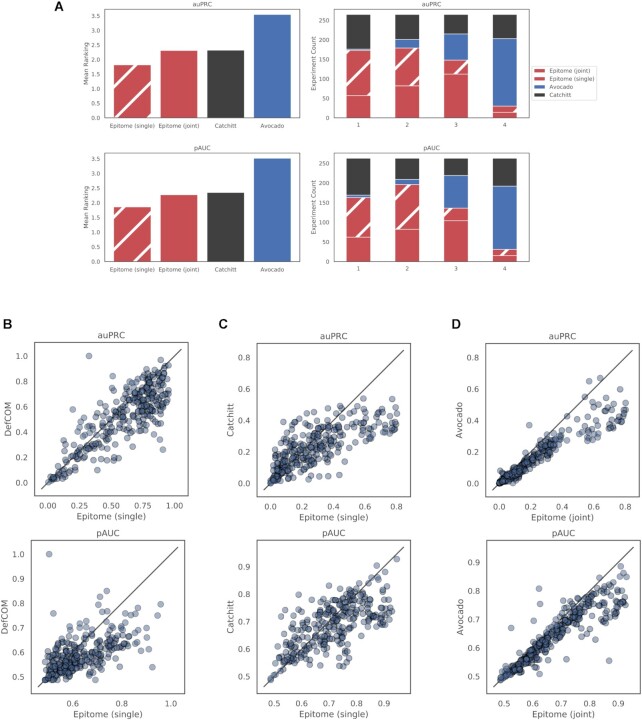
Figure 2.
Comparison of methods for predicting transcription factor binding sites (TFBS) for 77 transcription factors (TFs) and chromatin modifiers in 40 primary cells, cell lines, and tissues from ENCODE. Transcription factor binding sites were predicted on all 200 bp regions on chromosomes 8 and 9 that overlap at least one binding site in at least one of the 40 cell types considered. (A) Frequency at which each method obtains a rank for predicting TFBS across 77 transcription factors and chromatin modifiers in 40 held out cell lines, tissues, and primary cells, totaling 264 comparisons. Evaluated methods include Avocado (30), Catchitt (24), a joint Epitome model, and single Epitome models, where each TF is trained separately. (Left) Mean pAUC (5% FPR) and auPRC ranking for each method. (Right) Frequency at which each method obtains a rank based on pAUC and auPRC. (B) Scatter plots comparing auPRC and pAUC (5% FPR) between Epitome and DeFCoM. Only regions overlapping motifs specific to the TF being evaluated were considered. Both DeFCoM and Epitome trained individual models for each TF evaluated. (C) Scatter plots comparing auPRC and pAUC (5% FPR) between Epitome and Catchitt. Both Catchitt and Epitome trained individual models for each TF evaluated. (D) Scatter plots comparing auPRC and pAUC (5% FPR) between Epitome and Avocado. Both Avocado and Epitome trained joint models for all TFs evaluated.