Figure 2.
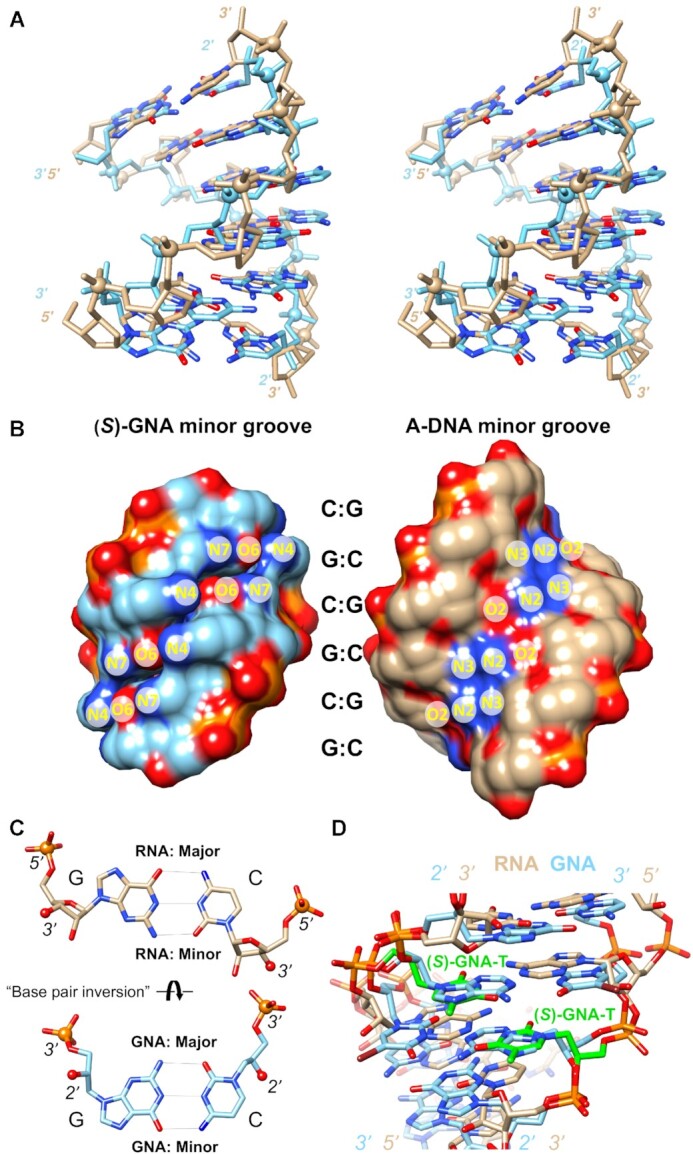
GNA displays an inverted base pair orientation relative to DNA and RNA. (A) Cross-eye stereo image of the (S)-GNA hexamer duplex (20) with sequence 3′-GCGCGC-2′ (light blue, PDB ID 2WNA) superimposed on the A-DNA model duplex with sequence 5′-GCGCGC-3′ (tan). Phosphorus atoms are highlighted as spheres and nucleobase oxygen and nitrogen atoms are colored in red and blue, respectively. The DNA duplex was built using the 3DNA program (38) and a 2.87 Å helical rise and a 33° twist. The overlay was done using phosphate groups and N1 (cytosine) and N9 (guanine) atoms, resulting in an r.m.s.d. of 2 Å. (B) The separated (S)-GNA and A-DNA duplexes from panel (A) in a surface rendering and viewed into the minor groove. Atom labels for guanine and cytosine H-bond donor and acceptor atoms indicate that base edges in the major and minor grooves are swapped in GNA relative to DNA and RNA. (C) Comparison between GNA and RNA G:C pairs that illustrates the rotation of base pairs around their long axis in the two systems, despite similar A-form backbone geometries and groove dimensions. RNA 5′-phosphorus and 3′-oxygen atoms as well as GNA 3′-phosphorus and 2′-oxygen atoms are drawn as spheres to highlight their similar relative orientations. (D) Overlay of the (S)-GNA octamer (19) duplex with sequence 3′-CTC-BrU-AGAG-2′ (PDB ID 2XC6) and the RNA duplex (21) of the sequence 5′-CGAATUCG-3′ with (S)-GNA-T (green carbon atoms) modification (PDB ID 5V2H). The view is into the minor groove and illustrates that the GNA thymidine retains its base-rotated orientation opposite RNA adenosine.
