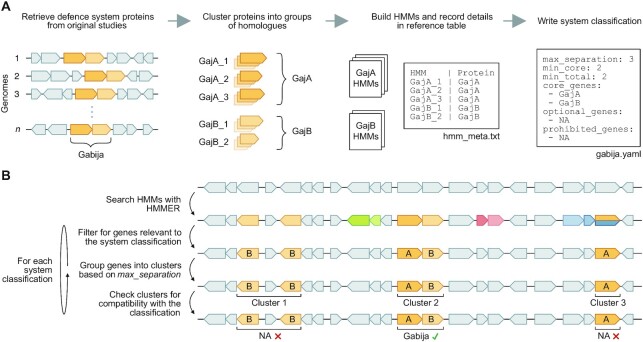
Figure 1.
Workflow of data preparation and PADLOC functioning. (A) Preparation of data for PADLOC. For each type of defence system protein, sequences were retrieved and clustered into homologue groups. An HMM was built from each group of proteins, and the names of the HMMs (e.g. GajA_1) and their corresponding protein families (e.g. GajA) were recorded in a reference table (hmm_meta.txt), which allows a single family of defence system proteins to be represented by multiple HMMs. A simple classification file ([system].yaml) was written to represent each defence system, describing the typical genetic architecture of the system. (B) Automated functional workflow of PADLOC. HMMER is used to identify genes encoding defence protein homologues in the input genome. Each system classification is then analysed individually, filtering the HMM hits for genes relevant to the current type of system being searched. HMM hits are grouped into gene clusters based on the synteny requirements specified in the system classification. Each cluster is then checked against the system classification to determine whether the system requirements are fulfilled. Yellow genes represent Gabija; green, red or blue genes represent genes from other defence systems; genes with two colours (i.e. yellow/blue) represent genes matched by HMMs from two different defence systems.