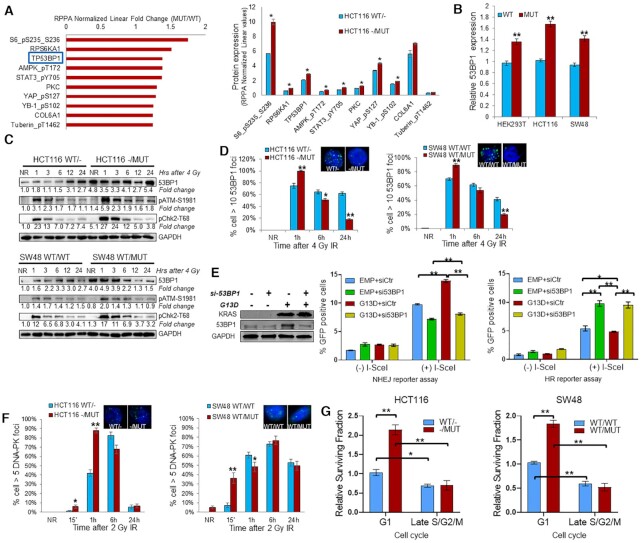
Figure 4.
KRAS mutation upregulates 53BP1 to influence DNA double strand break repair pathway choice. (A) TP53BP1 is one of the top 10 upregulated proteins in HCT116–/MUT relative to HCT116WT/- by RPPA analysis. Left panel: RPPA analysis displaying the fold change of top 10 upregulated protein expression or phosphorylation in KRAS mutant HCT116 cells relative to KRAS wild-type HCT116 cells. Right panel: RPPA analysis displaying the protein expression or phosphorylation levels in HCT116 isogenic cells. (B) Exponentially growing HCT116 and SW48 KRAS G13D isogenic cells, and HEK293T cells transfected with a plasmid expressing KRAS G13D or empty vector were subjected to qRT-PCR analysis of TP53BP1 mRNA. (C) Western blot and densitometric quantification of proteins at the indicated hours post 4 Gy IR (NR = no radiation, time zero hours). Fold change of protein are compared to KRAS wild-type cells without IR. (D) At the indicated hours post 4 Gy, isogenic HCT116 and SW48 cells were prepared for immunofluorescence of 53BP1 nuclear foci. Inset: representative pictures of foci at 24 h after IR. (E) HEK293T cells were transfected with KRAS G13D plasmid (G13D), empty vector, 53BP1 siRNA (si53BP1) or scrambled control siRNA (siCtr) for 48 h and subjected to immunoblotting. DNA damage reporter assays were performed by transfecting HEK293T cells with NHEJ-GFP or HR-GFP reporter plasmid, together with KRAS G13D, empty vector (EMP), si53BP1 or siCtr plasmid for 24 h, followed by addition of adenovirus containing I-SceI to induce DNA DSB in the reporter plasmid consensus sequence. After 24 h, the cells were analyzed for GFP-positive cells by flow cytometry to measure NHEJ or HR repair proficiency. The values represent the mean ± SD from triplicate dishes, and at least three independent experiments were performed. mCherry plasmid was used to normalize transfection efficiency. (F) At the indicated time post 4 Gy, isogenic HCT116 and SW48 cells were prepared for immunofluorescence of DNA-PKcs nuclear foci. Data shown as mean ± SD. Experiments were performed three times. (G) Clonogenic assay using cells enriched in G1 phase and late S/G2/M phase. Surviving fraction at 2 Gy was normalized to KRAS wild-type G1 cells; *P < 0.05, **P < 0.001