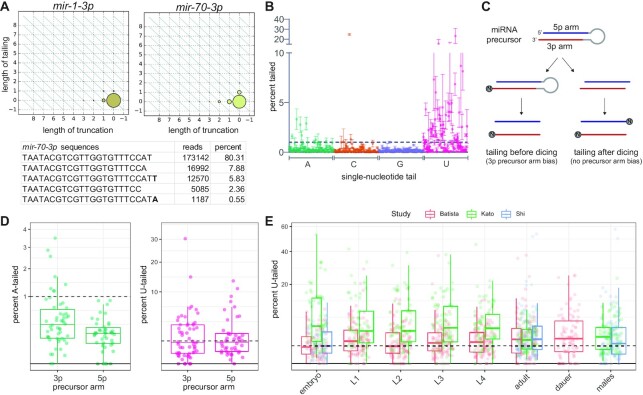
Figure 1.
Characterization of miRNA tailing in C. elegans. (A) 2D matrix representing the truncation and tailing status of the indicated miRNAs. The sizes of the dots represent the proportion of reads, with the canonical sequence at zero on both axes, and increasingly trimmed or tailed species plotted to the left on the x-axis or upward on the y-axis, respectively. Bottom: Number of reads of most abundant mir-70-3p species in one wild type adult library, with the canonical sequence being the most abundant. (B) Mean and standard deviation of prevalence of single nucleotide 3′ terminal additions of each indicated nucleotide from three biological replicates. (C) Schematic of miRNA tailing preceding or following Dicer-mediated cleavage of the miRNA precursor. (D) Comparison of prevalence of mono-adenylation and mono-uridylation on 3p- versus 5p-derived miRNAs. One biological replicate is shown. (E) Meta-analysis of three published datasets shows tailing across C. elegans developmental stages. Prevalence of mono-uridylation of miRNAs across development is shown. Only miRNAs with >50 RPM in the indicated library were analyzed. (B, D) Only miRNAs with >50 RPM in all biological replicates were analyzed. (B, D, E) Each dot represents an individual miRNA.