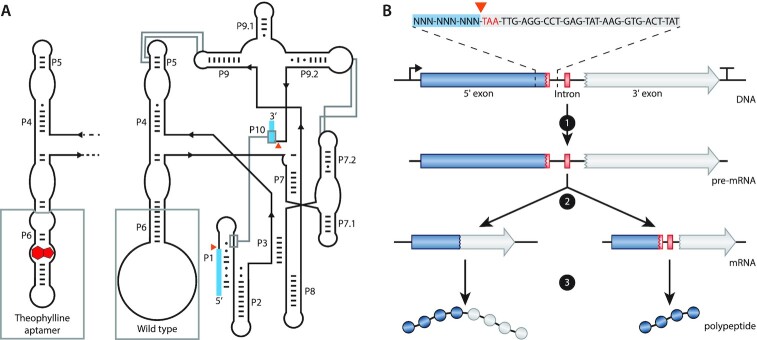
Figure 1.
Schematics and function of the T4 td intron. (A) Schematic representation of the predicted secondary and tertiary structure of the wild-type (WT; right) and the theophylline-dependent T4 td intron (left). The structure follows the format of Cech et al. (69). P1 to P10 represent the pairing domains of the intron. Exon sequences are indicated as blue boxes. Orange triangles indicate the splicing sites. Base pairs are indicated by ‘-’ and wobble pairs by ‘•’. The grey boxes at P6 highlight the difference between the WT and the theophylline-dependent ribozyme. Grey lines show interactions within the intron. (B) Schematic representation of the transcription and translation of a gene containing the T4 td intron in its open reading frame. At the top, the intron in-frame stop codon (TAA) is depicted in red, the 5′ flanking region is highlighted with a blue box and a part of the intron sequence is highlighted with a light grey box. 1 depicts transcription, 2 depicts self-splicing (left path) or no self-splicing of the intron (right path) and 3 depicts translation of the full protein (left path) or the translation of a truncated protein (right path) when the intron is spliced or retained, respectively.