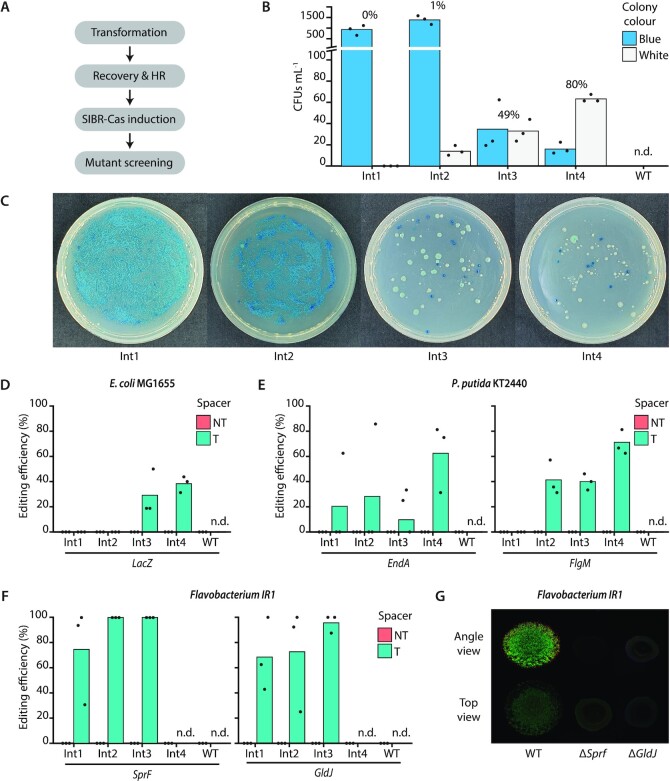
Figure 4.
SIBR-Cas genome editing assays in E. coli MG1655, P. putida KT2440 and Flavobacterium IR1. (A) Schematic for SIBR-Cas editing procedure. The time required for recovery and HR differs amongst the different bacterial species as described in the materials and methods section. (B) Editing efficiency of the LacZ gene in E. coli MG1655. Blue/white screening was performed to distinguish the edited (white) from the unedited (blue) colonies when using either of the four different SIBR-Cas variants Int1 (0%), Int2 (1% ± 0.35%), Int3 (49% ± 8.75%) and Int4 (80% ± 5.57%) or the WT-FnCas12a (0%, n.d.). The percentage on top of each variant indicates the percentage of white colonies from the total number of colony forming units mL–1 (CFUs ml–1). (C) Representative plates of edited E. coli MG1655 cells at the LacZ locus using the four different SIBR-Cas variants (Int1-4; Int1 is the worst and Int4 is the best splicer). (D) Unbiased (omitting the presence of X-Gal in the medium) editing efficiency of the LacZ gene using the four different SIBR-Cas variants Int1 (0%), Int2 (0%), Int3 (29% ± 18.04%), Int4 (38% ± 6.41%) or the WT-FnCas12a (0%, n.d.). All of the NT controls showed 0% targeting efficiency. (E) Editing efficiency of the EndA [Int1 (21% ± 36.08), Int2 (29% ± 49.49%), Int3 (19% ± 17.35%), Int4 (63% ± 27.24%) or the WT-FnCas12a (0%, n.d.)] and FlgM [Int1 (0%), Int2 (41% ± 13.84%), Int3 (40% ± 6.41%), Int4 (70% ± 9.85%) or the WT-FnCas12a (0%, n.d.)] genes in P. putida KT2440. All of the NT controls showed 0% targeting efficiency. (F) Editing efficiency of the SprF [Int1 (75% ± 38.29), Int2 (100% ± 0%), Int3 (100% ± 0%), Int4 (0%, n.d.) or the WT-FnCas12a (0%, n.d.)] and GldJ [Int1 (68% ± 29.03%), Int2 (72% ± 41.26%), Int3 (96% ± 7.22%), Int4 (0%, n.d.) or the WT-FnCas12a (0%, n.d.)] genes in Flavobacterium IR1. Individual bars represent the mean of triplicate experiments and ‘•’ represents the value of each replicate. N.d., not determined. All of the NT controls showed 0% targeting efficiency. (G) Comparison between WT Flavobacterium IR1 and ΔSprF and ΔGldJ strains generated with SIBR-Cas. Images were taken after incubation at room temperature for 2 days by inoculating 3 μl spot on ASWBC solid medium (ASW supplemented with black ink and carrageenan). The WT strain (left) is 18 mm across.