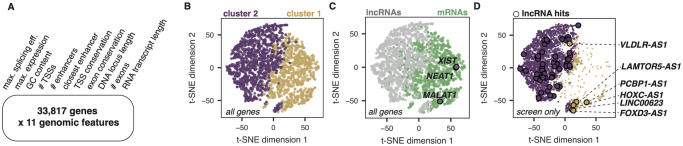
Fig 6. Using k-means clustering of 11 genomic features to find candidate functional RNA molecules.
(A) List of the 11 genomic features used in clustering analysis across 33,817 genes. (B) t-SNE visualization of the data matrix outlined in (A), colored by k-means cluster assignment. Plot shows a sub-sample of the data: 1000 randomly-sampled lncRNAs and 1000 randomly-sampled mRNAs. (C) Same t-SNE as in (A), now colored by biotype, where gray dots are lncRNAs and green dots are mRNAs. The 3 “gold standard” lncRNAs (XIST, NEAT1, and MALAT1) are highlighted and outlined in black. (D) Subset of the same t-SNE in (A) and (B), now showing only lncRNAs included in the screen, colored by their cluster assignment. Significant hits in either cluster are outlined in black. Hits assigned to cluster 1 are annotated.