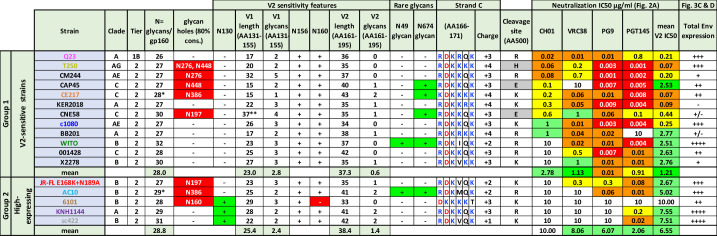
Fig 1. Key features of candidate Env strains.
17 strains were split into group 1 strains (n = 12) that are naturally sensitive to multiple V2 NAbs and group 2 strains (n = 5) that exhibit high membrane trimer expression. Strain names are abbreviated (see Materials and Methods). An asterisk in total glycans/gp160 protomer indicates overlapping sequons in the CE217 and AC10 strains, only one of which can carry a glycan. Glycan holes are listed whenever a ≥80% conserved glycan is absent. V2 sensitivity features are shown, including glycans involved in NAb binding or clashes and loop lengths. A double asterisk for the CNE58 V1 loop denotes a possible internal hairpin disulfide loop (S1 Fig). Rare glycans N49 and N674 are shown. Strand C sequence (AA166-171) is shown with basic residues in blue and acidic residues in red, along with its charge. The residue at position 500 may influence gp120/gp41 processing (gray highlights non-lysine or arginine residues). PV IC50s for V2 NAbs (CF2 assay) (see Fig 2A). JR-FL neutralization data is for the E168K+N189A mutant. Total Env expression, as judged by SDS-PAGE-Western blot (Fig 3C and 3D).