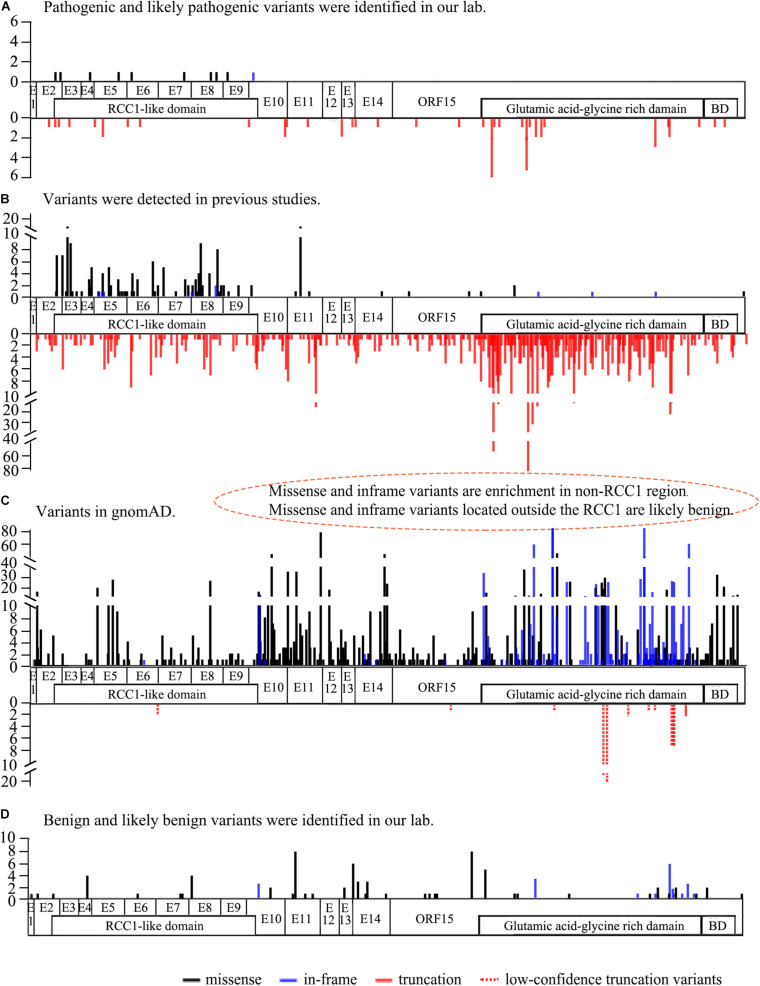
FIGURE 1.
The frequency and location of the variants from our lab, previous studies, and the gnomAD database (Ref. NM_001034853). (A) The frequency and location of pathogenic and likely pathogenic RPGR variants detected in our lab. Missense and in-frame variants are distributed above the structure, and truncation variants are shown below the structure. (B) The frequency and location of RPGR variants identified in previous studies. Missense and in-frame variants enriched in the RCC1-like domain are shown above the structure, and truncation variants are indicated below the structure. Gross deletion variants are not shown here. (C) The frequency and location of RPGR variants from the gnomAD database. Missense and in-frame variants are significantly enriched in the non-RCC1-like domain above the structure. Truncation variants in all coding regions below the structure. Of the 11 truncation variants, 10 were low confidence truncations (dotted line). (D) The frequency and location of benign and likely benign RPGR variants identified in our lab. The white regions represent the coding regions. RCC1-like domain: p.38∼367, BD: basic domain p.1086-1139, Glutamic acid-glycine-rich domain: p.728∼1084.