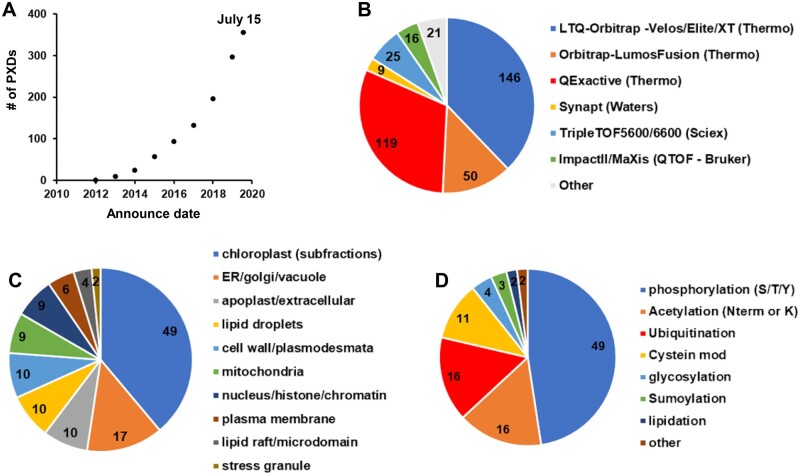
Figure 2.
Features of PXDs for Arabidopsis available via ProteomeXchange through July 15, 2020. Information about these PXDs was obtained from the submitted metadata and/or accompanying publications. A, Accumulative PXDs with verified Arabidopsis content by year (2010-7/2020). B, Type of MS Instrument (LTQ-Orbitrap–Velos/Elite/XT [Thermo], Orbitrap-LumosFusion [Thermo], QExactive [Thermo], Synapt [Waters], TripleTOF5600/6600 [Sciex], ImpactII/MaXis [Bruker], other). C, Arabidopsis subcellular fractions (plastid, mitochondria, peroxisomes, vacuole, nucleus, apoplast/extracellular, cytosol, ER/Golgi/PM). D, Post-translational modifications that were specifically enriched prior to MS analysis (phosphorylation, acetylation [N-term or Lys], ubiquitination, cysteine oxidation, glycosylation, sumoylation, lipidation, other).