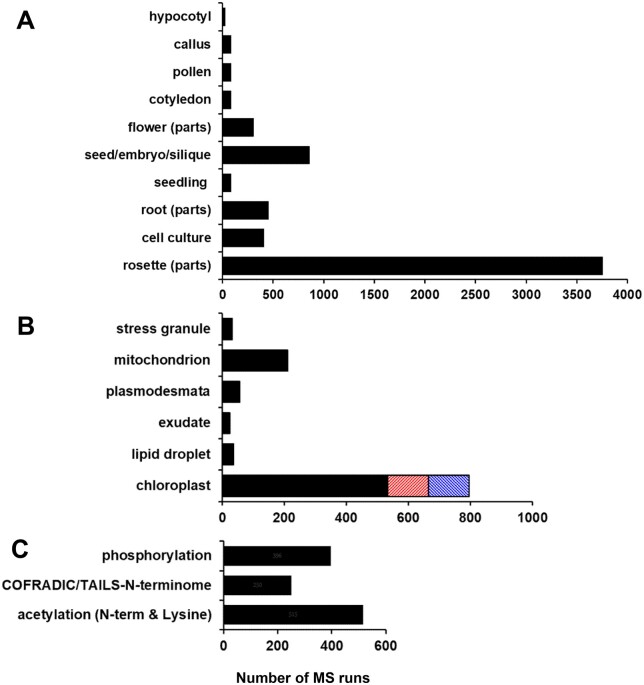
Figure 3.
Key features of the samples used for the raw files (MS runs) for the 52 selected PXDs for the first Arabidopsis PeptideAtlas build. The count is based on the number of MS runs (raw files) for each part. A, Arabidopsis plant parts—hypocotyl, callus, pollen, cotyledon, flower parts (sepal/petal/carpel/stamen/pedicle, seed/septum/embryo), seedling (all parts of a young plant—root-hypocotyl/cotyledons/few young leaves, mostly collected from plates or liquid culture), root (tip/exudate/zone), cell culture, rosette parts (rosette/leaf/petiole/cauline leaf/senescing leaf/stem/internode). B, Arabidopsis subcellular fractions specifically analyzed are stress granule, mitochondrion, plasmodesmata, root exudate, cytosolic lipid droplet, chloroplast (black), and the specific fractions thylakoid (orange), plastoglobuli (blue). C, MS runs of samples that were specifically prepared to analyze PTMs (phosphorylation, acetylation of the N-terminus, and/or lysine) or to determine the physiological N-terminus using N-terminome enrichment techniques (TAILS, COFRADIC, or ChaFRADIC).