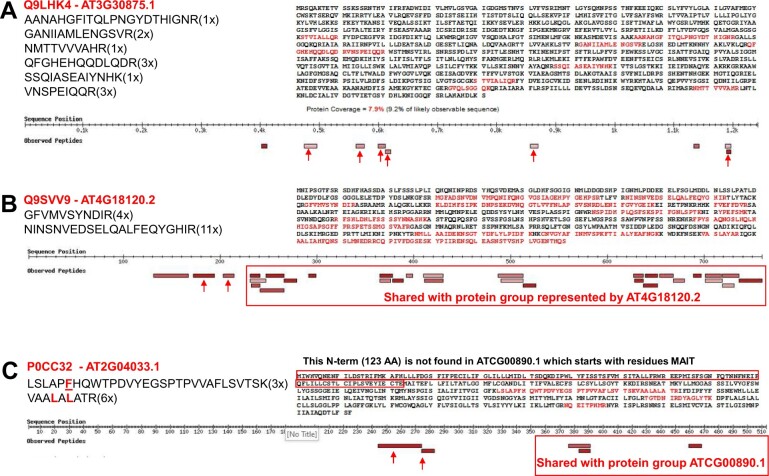
Figure 9.
Examples of UniProtKB identifications selected from the 49 identifications based on at least two distinct peptides (not matched to predicted Araport11 proteins) that were observed at least three times. For a complete listing, see Supplemental Data Set S3D. The examples were selected because the UniProtKB protein sequence matched with relative high significance to a predicted pseudogene in Araport11 (based on TBlastN). A, Q9LHK4 encodes a large 137-kDa protein (1,241 aa). TBlastN identified Araport11 AT3G30875 as a strong match (1,215 aa alignment length and E = 0). AT3G30875 is annotated as a pseudogene (nine exons are shown in Jbrowse), but the TAIR website notes that this is probably not a pseudogene based on evidence for transcription (RNA-seq) and translation (Ribo-seq) described in Hsu et al. (2016). We identified six peptides (marked with red arrows) uniquely mapping to Q9LHK4 (sequences are shown), two of which were observed three times, and the others only once or twice. An additional three matched peptides were shared with other proteins in the protein group represented by AT4G17140.3, which is a protein with repeating coiled regions of VPS13. B, Q9SVV9 encodes an 85 kDa protein (759 aa) and is identical to TAIR10 AT4G18120.2, with the exception of two gaps in the amino acid sequence alignment. This protein (AML3) is encoded by a member of the MEI2-like gene family and is annotated as a pseudogene in Araporti1. In situ hybridization detected expression during early embryo development but not in vegetative or floral apices (Kaur et al., 2006). Two peptides uniquely map to Q9SVV9 as indicated by the red arrows. The other matched peptides are shared with the protein group represented by AT4G18120.2 as indicated. C, P0CC32 encodes a 57-kDa protein (512 aa) and maps to the pseudogene AT2G04033.1 with similarity to the defensin-like (DEFL) family. However, careful inspection of the results for P0CC32 in PeptideAtlas shows that the Uniprot sequences is nearly identical to a much smaller (42 kDa) chloroplast-encoded NDHB/NDH1 protein (ATCG00980.1), with the exception of an N-terminal region of 123 aa. P0CC32 was identified as having two unique peptides that did not match to AT2G04033.1 because this protein is RNA edited, and these two peptides contain one or two editing sites resulting into amino acid changes. As mentioned in the “Results and discussion”, the Araport11 sequences are in the unedited form, whereas UniProtKB does incorporate these edits. Three additional peptides were identified for P0CC32, but these were all shared with AT2G0433.1, as indicated.