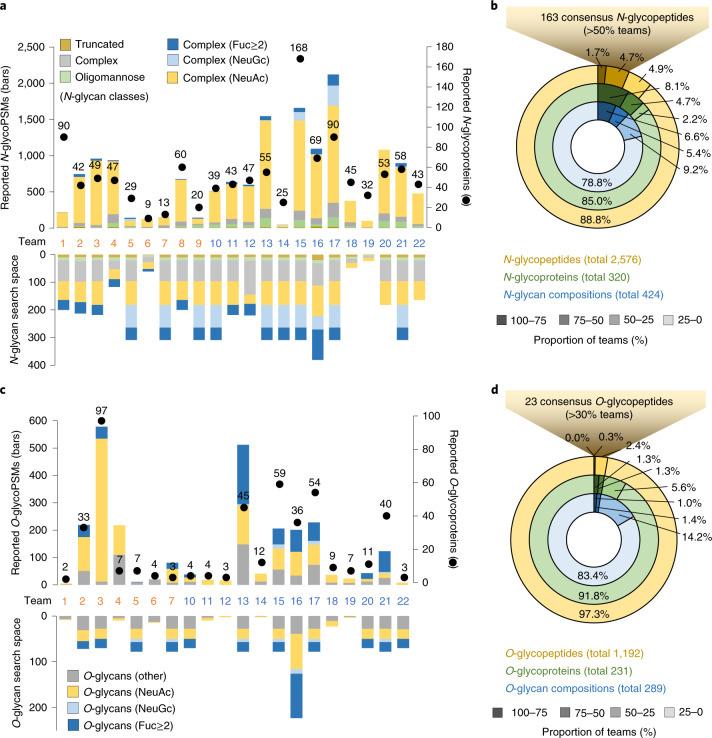
Fig. 2. Glycopeptides reported across teams.
a, Reported N-glycoPSMs (bars), unique source N-glycoproteins (dots) and the N-glycan search space applied (mirror bars) by each team. See key for N-glycan classification. b, Proportion of N-glycopeptides, source N-glycoproteins and N-glycan compositions commonly reported by teams. c, Reported O-glycoPSMs, unique source O-glycoproteins and the O-glycan search space applied by each team. Teams 8 and 9 did not perform O-glycopeptide analysis. See key for O-glycan classification. Multi-feature N- and O-glycans fitting into several of these classes were for this purpose classified in a prioritized order of multi-Fuc–NeuGc–NeuAc; see Supplementary Tables 2 and 3 for data. d, Proportion of O-glycopeptides, source O-glycoproteins and O-glycan compositions commonly reported by teams. The high-confidence ‘consensus’ N- and O-glycopeptides have been made publicly available (GlyConnect Reference ID 2943).