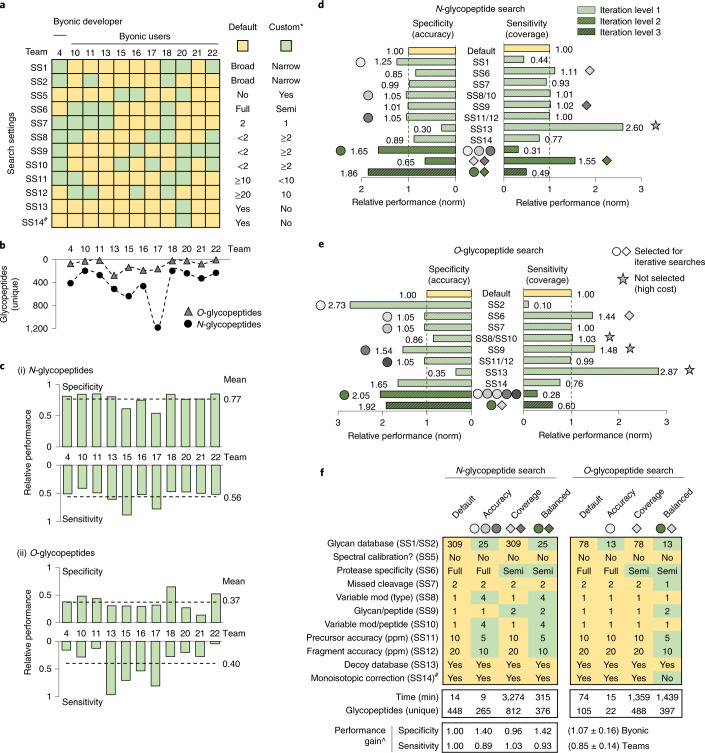
Fig. 4. Search engine-centric (Byonic-focused) analysis of search strategies for high-performance glycoproteomics data analysis.
a, Overview of the search settings employed by Byonic teams. Default: search strategy used by most teams (yellow). Custom: variations from the default search strategy (green). #Data for SS14, a setting not included in the team reports, were adopted from SO4 data. b, The glycoproteome coverage (unique glycopeptides, File B) varied among Byonic teams. c, Specificity (accuracy) and sensitivity (coverage) scores for (i) N-glycopeptides and (ii) O-glycopeptides for Byonic teams. d, Controlled (in-house) searches for N-glycopeptides using Byonic (File B). Individual search settings were systematically varied (iteration level 1) and output assessed for performance gains (specificity, sensitivity). Search settings showing performance gains (shaded circles/diamonds) without unacceptable costs in specificity (SS13) or search time (SS8/SS10, SS9; see examples in e) (gray stars) were collectively tested for synergistic performance gains (iteration levels 2 and 3, dark green). See e for shared symbol key. e, Byonic-centric O-glycopeptide searches. See d for details. f, Recommended Byonic-centric search strategies for ‘high accuracy’, ‘high coverage’ and ‘balanced’ (between accuracy and coverage) glycoproteomics data analysis. ^The recommended search strategies showed relative performance gains as determined using an independent glycoprotein-centric score (Supplementary Table 19b). Search time and glycoproteome coverage (unique glycopeptides) are also indicated.