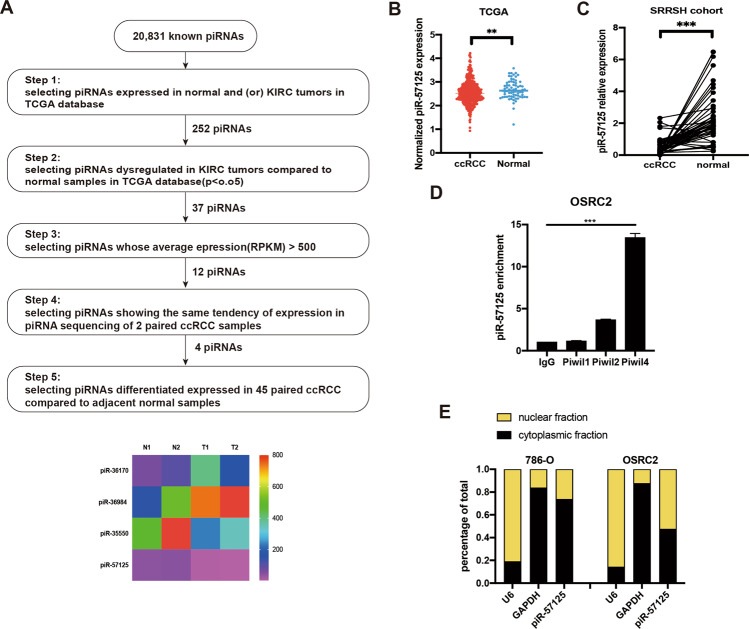
Fig. 1. piR-57125 is downregulated in ccRCC.
a Top, flow chart demonstrates the selection criteria of potential piRNAs dysregulated in ccRCC. Bottom, heatmap showing the differential expression of piRNAs in 2 pairs of ccRCC and adjacent normal samples by piRNA sequencing. b Expression of piR-57125 in ccRCC and normal patients based on the analysis of TCGA data. c qRT-PCR analysis of piR-57125 expression in 45 paired ccRCC and adjacent normal samples from patients recruited from the SRRSH cohort. Data represent mean ± S.D. d RIP and RT-qPCR assay showed that PIWIL4 has more affinity to piR-57125 in OSRC-2 cells compared with PIWIL1 and PIWIL2. e Cytoplasm and nuclear mRNA fractionation experiment showed that piR-57125 exists in both the cytoplasm and the nucleus. U6 and GAPDH act as the positive controls of nucleus and cytoplasm, respectively. **p < 0.01; ***p < 0.001.