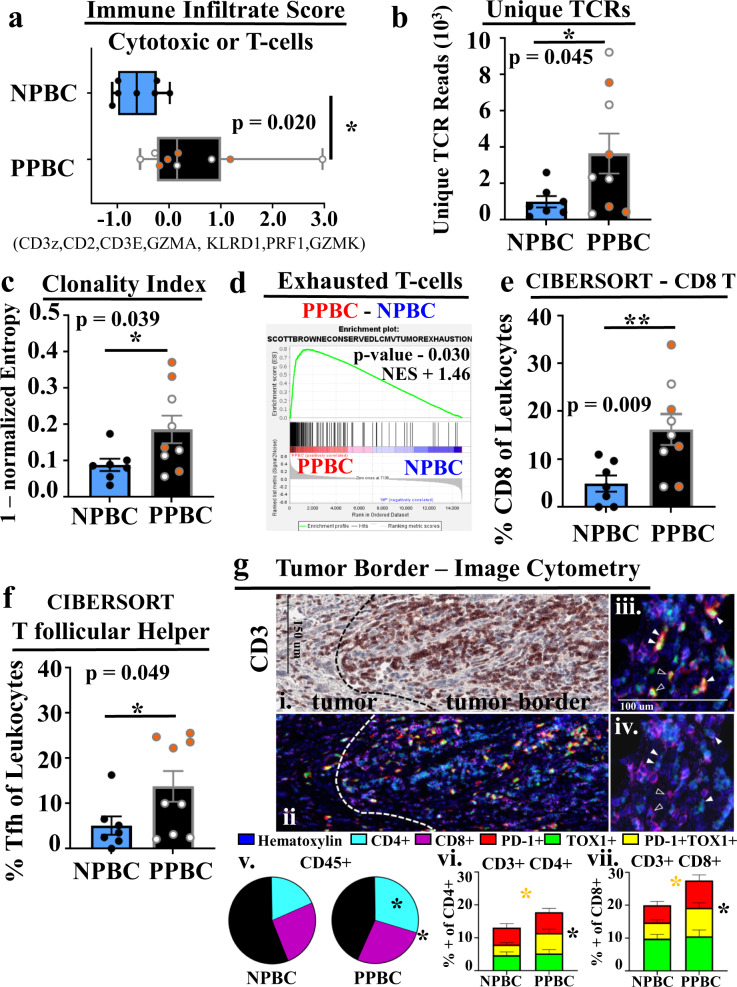
Fig. 4. PPBC is enriched for activated T-cells compared with NPBC.
Characterization of the immune cell presence and T-cell immunity enriched in postpartum breast cancer (PPBC) compared with nulliparous breast cancer (NPBC). a RNA Seq expression data was evaluated for genes associated with cytotoxic or T-cell immunity using a gene signature called the single sample immune infiltrate score (PPBC n = 9, NPBC n = 7). b Unique numbers of T-cell receptors (TCR) for each RNA Seq sample, compared between groups (PPBC n = 9, NPBC n = 7). Relative clonality demonstrated by c normalized (norm) entropy (PPBC n = 9, NPBC n = 7). d GSEA profile-derived from exhausted T-cell signature. CIBERSORT deconvolution of RNA Seq data depicts increased e CD8 T-cell and f T follicular helper (Tfh) presence as a fraction of total leukocytes (PPBC = 9, NPBC = 7). g multiplex IHC analysis of the tumor border in NPBC (n = 13) and PPBC (n = 14) cases was subjected to quantification by image cytometry. gi Hematoxylin (blue) stain & AMEC (red/brown) for CD3 demonstrating T-cell accumulation in the tumor border region. Dashed lines indicate demarcation of intratumoral and tumor border regions. gii Aligned pseudo colored multiplex IHC images depicting staining from hematoxylin (dark blue) and chromagen mediated antibody detection of CD4+ (light blue) CD8+ (purple), PD-1+ (red) or TOX1+ (green) cells. PD-1+/TOX1+ cells giii appear yellow due to overlap of red and green coloring. giv PD-1+ and TOX1+ cells depicted in giii can be either CD4+ (white arrows) or CD8+ (black arrows) T-cells. gv Pie-charts depicting increased CD4+ CD3+ T-cells (light blue, p = 0.0052) and total T-cell content (light blue and purple, p = 0.0225) as fraction of CD45+ cells in the PPBC cohort. PD-1+ (red), TOX1+ (green) or PD-1+ TOX1+ (yellow) cells as a fraction of the gvi CD45+ CD3+ CD4+ (yellow group comparison = yellow star p = 0.0225, PD-1/red + yellow comparison = black star p = 0.05) or gvii CD45+ CD3+ CD8+ (yellow group comparison = yellow star p = 0.0205, PD-1/red + yellow comparison = black star p = 0.028) T-cell compartments. P values determined by GSEA software (nominal, non-adjusted p value, d) or Students’ unpaired two-tailed t test with Welch correction (a–c, e, f), or Students’ unpaired one-tailed t test with Welch correction for confirmatory IHC(*p ≤ 0.05, g). ICGC identified TP53 mutations are noted by orange-filled circles. For box and whisker plot (a) data are presented as minimum to maximum with median value marked by a line within the depicted interquartile range, whereas data depicted in bar graphs (b, c, e, f, gvi, gvii) are presented as mean values ±SEM.