Figure 6.
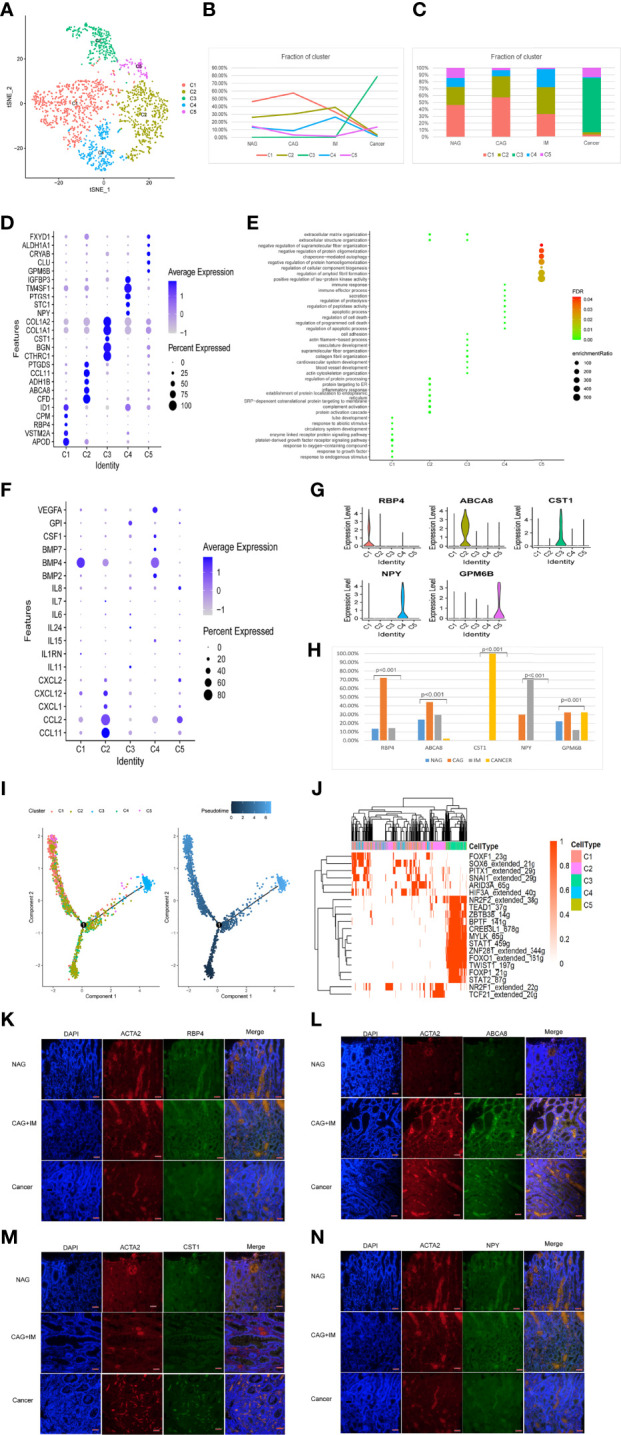
Identification of fibroblast clusters and expression features at different stages. (A) tSNE plot of five fibroblast cell subclusters. (B) Line chart displaying changing trend of the proportion of the five cell types across the four stages. (C) Stacked histogram showing fibroblast composition across the four stages. (D) Bubble plot of the top five markers of each cell cluster; dot sizes represent abundance while colors represent expression levels. (E) Bubble plot showing the biological functions of different cell clusters using GO; dot sizes represent abundance while colors represent q values. (G) Violin plots of marker genes in the five subclusters. (F) Bubble plot showing scale normalized expression of representative genes involved in chemokine, cytokine, and growth factor processes. (H) Histograms of scale normalized expression levels of three marker genes at each stage. (I) Fibroblast cell differentiation trajectory with each color coded for pseudotime (right) and clusters (left). (J) AUC scores of transcription factor expression regulation using SCENIC. Results converted to binary data were visualized as heatmap plots constructed using the pheatmap function of R. (K) Immunofluorescence staining of RBP4 (green) and ACTA2 (red) in different stages of stomach disease. Scale bars, 50 μm. (L) Immunofluorescence staining of ABCA8 (green) and ACTA2 (red) in different stages of stomach disease. Scale bars, 50 μm. (M) Immunofluorescence staining of CST1 (green) and ACTA2 (red) in different stages of stomach disease. Scale bars, 50 μm. (N) Immunofluorescence staining of NPY (green) and ACTA2 (red) in different stages of stomach disease. Scale bars, 50 μm.
