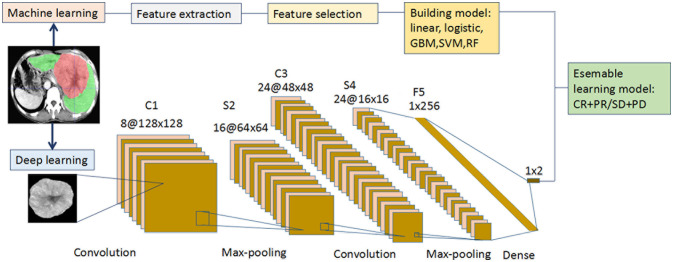
Figure 1.
Flowchart for the integration of the machine learning and deep learning models. (i) The radiologists manually segmented the 3D ROIs of HCC using ITK-SNAP. Thereafter, 1167 features were extracted from the hepatic arterial CT images based on the “pyradiomics” package of python. Redundant radiomics features were then eliminated by ICCs. Using RFE and 5-fold cross-validation, the final features were selected and the five radiomics models were built using different machine learning algorithms. (ii) A CT image and mask of ROI manually segmented from the largest tumor layer was resized and output as 224 × 224 ×3 from each patient in the two cohorts. Augmentation techniques were used to enlarge the training CT image dataset, and “new” big data were generated. The deep learning framework included two convolutions, two max-poolings, and one dense layer. The final output layer was a softmax classifier. The optimizer was Adam, with a learning rate of 0.001 and batch size of 16. All the layers were standardized and the L2 regularization was set to 0.000001. The activation function of RELU was set as alpha = 0.1. (iii Each of the five machine learning models (linear, logistic, GBM, SVM, and RF) was integrated with the deep learning model to predict the initial treatment response to TACE. CT, computed tomography; GBM, gradient boosting machine; HCC, hepatocellular carcinoma; ICC, intraclass correlation coefficient; RELU, reconstructed linear units; RF, random forest; RFE, recursive feature elimination; ROI, region of interest; SVM, support vector machine; TACE, transarterial chemoembolization.