FIGURE 1.
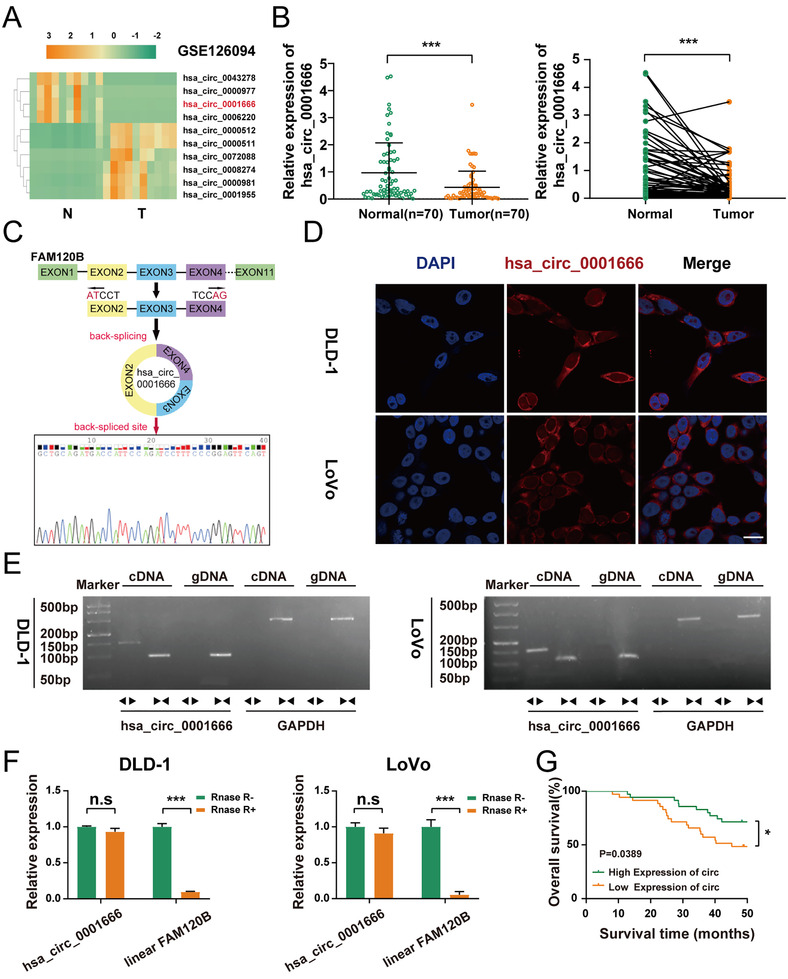
Hsa_circ_0001666 is lowly expressed in CRC tissues and cell lines and mainly localized in the cytoplasm. (A) Heatmap based on GSE126094, four most downregulated and six most upregulated circRNAs (adjusted p<.05 and log|FC|>4). (B) qRT‐PCR was used to detect the relative expression level of hsa_circ_0001666 in CRC tissues and matched para‐cancer tissues (n = 70). (C) The schematic illustration showed the FAM120B exon two to four circularization into hsa_circ_0001666; Sanger sequencing showed the head‐to‐tail splicing of hsa_circ_0001666. (D) The FISH assay revealed that hsa_circ_0001666 was mainly found in the cytoplasm. The nuclei were dyed blue with DAPI, and the hsa_circ_0001666 was stained red (scale bar, 100 μm; magnification, 600×). (E) RT‐PCR confirmed the existence of hsa_circ_0001666 both in DLD‐1 and LoVo. Divergent primers amplified hsa_circ_0001666 in cDNA but not in gDNA. GAPDH was used as a negative control. (F) The expression of hsa_circ_0001666 and FAM120B mRNA in both DLD‐1 and LoVo cell lines was detected by qRT‐PCR with or without RNase R. (G) Kaplan–Meier survival curves of CRC patients with low and high hsa_circ_0001666 expression. The median value was used as a cutoff. Data were all showed as mean ± SD (n = 3); n.s indicated no significance, *p < .05, ***p < .001
