Figure 4.
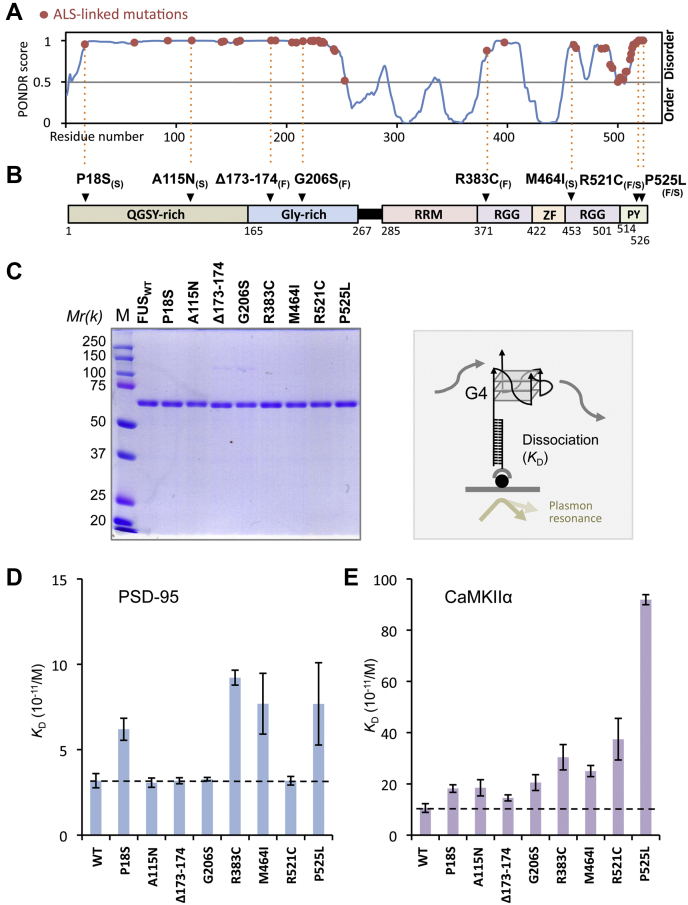
G4-RNA-binding kinetics of eight FUS mutants.A, order/disorder level was calculated for FUS using PONDR algorithm (Molecular Kinetics, Washington State University). The positions of ALS-linked 56 missense and internal deletion/insertion mutations are shown along the FUS protein from N- to C-terminus (40, 41, 42). B, a total of eight mutants with ALS-linked familial(F), sporadic(S), or both(F/S) at various locations on the FUS gene were selected and analyzed in this study. The position of each mutation is shown along the FUS map in (A). C, full-length mutant FUS proteins were purified in soluble forms according to the same procedure we developed for wild-type (31). One microgram each of the purified FUS proteins was analyzed by 10% SDS-PAGE, and the gel was stained with Coomassie brilliant blue. Right panel shows a schematic diagram of the SPR assay system. Terminal-biotinylated poly dT16 was bound to the streptavidin-coated sensor chip. Poly-dA16 tailed G4-RNA (20 nM) was immobilized onto the sensor chip, and various concentrations of wild-type and mutant FUS proteins were injected as the analyte. In this method, the dT16 oligomer was immobilized onto flow cell-2, and flow cell-1 was left blank to serve as in-line reference surface, RNA and analyte was injected to the flow cells-1 and cell-2 of the sensor chip. Plasmon resonance values (resonance unit; RU) were obtained from the flow-cell-2 data after subtracting the flow-cell-1 data. D and E, the dissociation constants (KD) of wild-type and mutant FUS proteins with PSD-95 or CaMKIIα G4-RNA. The experiments were performed three times, and the y-axis represents the mean ±SEM value. Kinetic data of the sensorgram are shown in Figures S4 and S5.