Figure 3. RBR‐type E3 ligases involved in coating cytosolic bacteria with ubiquitin.
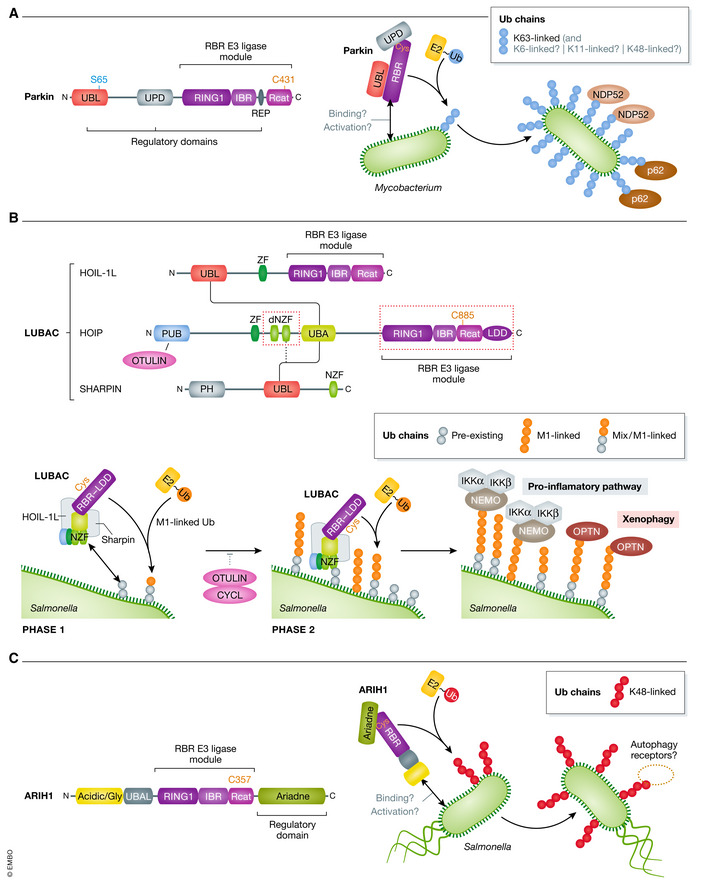
(A) Diagram of Parkin showing the Ub‐like (UBL) domain, the unique parkin domain (UPD), and the repressor element (REP), which form the regulatory part of Parkin, and the catalytic part comprising the RBR module at the C terminus. The mechanism for activation of Parkin in response to bacterial infection remains to be addressed but upon Mycobacterium infection, activated Parkin decorates the cytosolic bacteria with an Ub coat (predominantly K63 chains) that recruits p62 and NDP52 autophagy adaptors triggering xenophagy. (B) Domain organization of the three subunits of LUBAC: HOIP, HOIL‐IL, and SHARPIN. HOIL‐IL and SHARPIN interact via their respective UBL domain with the UBA domain of HOIP (solid lines), with potential contribution of the double NZF (dNZF) domain (dotted line). The dNZF and RBR domains of HOIP (red dotted boxes) are required for binding Salmonella in a two‐phase mechanism: in the first phase, LUBAC binds to preexisting Ub chains on Salmonella, in the second phase, enhanced HOIP activity mediates M1‐ and mixed M1‐linked Ub chains synthesis on these preexisting chains and triggers further LUBAC binding. The M1‐Ub chains recruit adapter proteins OPTN and NEMO which initiate xenophagy and pro‐inflammatory response, respectively. DUB activity of OTULIN and CYLD may counterbalance LUBAC activity. (C) Schematic view of ARIH1 domains showing the acidic/glycine region, UBA‐like domain (UBAL), the RBR module, and the regulatory Ariadne domain at the C terminus. ARIH1 decorates the Salmonella with K48‐Ub chains. The mechanism of its activation, binding to the cytosolic Salmonella, and the downstream action of ARIH1‐mediated K48‐Ub chains are not understood.
